PCR is a widely used technique in molecular biology, but its performance can be significantly impacted by PCR inhibitors. Understanding what PCR inhibitors are, how they affect amplification, and how to remove them is key to achieving reliable results.
What is PCR?
Polymerase chain reaction (PCR) is a molecular biology technique in which specific regions of DNA are amplified in vitro. It is one of the most widely utilised methods in life sciences due to its broad applicability in downstream molecular workflows. These include pathogen surveillance, cloning, in vitro diagnostics, DNA fingerprinting and genotyping.¹
PCR is also a critical step in targeted Next Generation Sequencing (NGS), as it enables enrichment of specific DNA fragments before sequencing. Several types of PCR exist, such as reverse transcription-polymerase chain reaction (RT-PCR) and quantitative polymerase chain reaction (qPCR). The specific approach depends on the nucleic acid template and whether quantification is required.
All PCR types require a mix of DNA or RNA template, a polymerase enzyme, a primer pair, nucleotide bases, and co-factors such as magnesium ions. This reaction mixture is processed in a thermocycler, where temperature cycling enables DNA denaturation, primer annealing, and strand elongation (Figure 1).
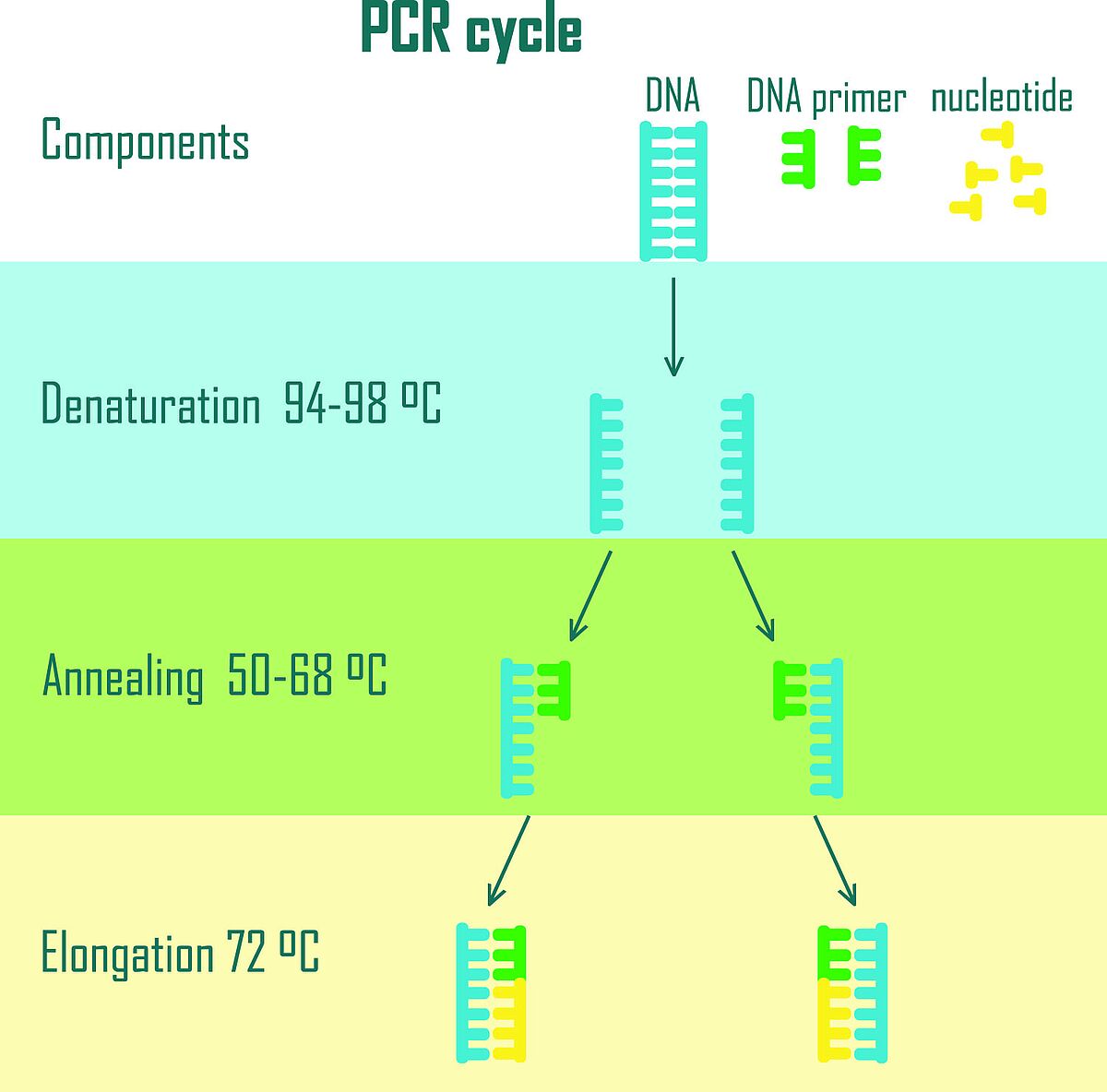
As robust and essential as PCR is, its high sensitivity also makes it vulnerable to various sources of error. A common challenge is interference by PCR inhibitors.
What are PCR inhibitors?
PCR inhibitors are organic or inorganic substances that interfere with the amplification reaction.¹ These can include simple salts (e.g. sodium, calcium) or more complex compounds such as polyphenolics, polysaccharides, detergents, or EDTA.
These inhibitors can enter the sample during preparation or be co-purified with DNA or RNA during extraction. Environmental sources such as soil, wastewater, faeces, or plant material are often rich in inhibitory substances like tannins or humic acids.¹ ² Biological samples such as blood or tissue may contain melanin, collagen, or haematin, while textile samples can carry inhibitory dyes like indigo.¹
PCR inhibitors may affect amplification in different ways. Some bind to the DNA polymerase, reducing or halting strand elongation. Others bind the DNA template, interfering with denaturation.³ Certain molecules like tannins or EDTA can chelate essential cofactors (e.g. Mg²⁺), decreasing the enzyme’s activity or completely inactivating it.³
Are PCR inhibitors affecting your data?
A key concern with PCR inhibitors is the risk of false-negative results. If the target DNA is not amplified due to inhibition, it may appear to be absent even when present.
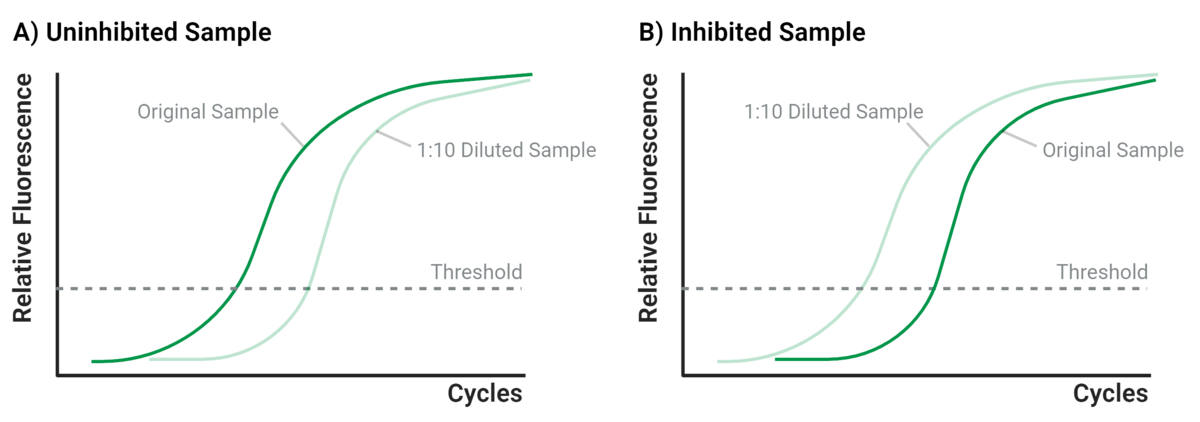
A practical method to test for inhibition is sample dilution. This reduces inhibitor concentration, potentially restoring amplification (Figure 2). In real-time qPCR, results are typically interpreted based on the Ct (cycle threshold) value – the number of cycles needed for fluorescence to exceed background levels. Lower Ct values indicate higher starting template amounts.
In Figure 2a, an uninhibited 1:10 diluted sample shows a higher Ct than the undiluted sample. In Figure 2b, the Ct remains equal or lower in the diluted sample, indicating inhibition in the undiluted one.
How to remove PCR inhibitors
There are multiple strategies to reduce the effects of PCR inhibitors. Increasing the concentration of DNA polymerase or cofactors may compensate for inhibition. However, this is not always sufficient.
Dilution is another common method, but it may reduce assay sensitivity. Conventional purification techniques include phenol-chloroform extraction, ion-exchange chromatography, cation exchange columns, or CTAB-based protocols.¹ While effective, these are often time-consuming, labour-intensive, and may involve hazardous chemicals. Repeated purification steps can also result in significant nucleic acid loss.
To address this, spin-column based purification systems have become widely adopted. Zymo Research offers products such as the DNA Clean & Concentrator-5, which effectively removes inhibitors like salts and detergents. However, for more complex inhibitors like polyphenolics, co-purification remains an issue.
Zymo Research’s OneStep PCR Inhibitor Removal Kit is designed to overcome this challenge by removing polyphenolic PCR inhibitors from DNA and RNA in under 5 minutes – without harsh chemicals. The kit uses a proprietary column matrix that binds substances like tannins, melanin, and humic/fulvic acids. A single centrifugation step minimises nucleic acid loss. This technology is also integrated into Zymo’s extraction kits, including the Quick-DNA Fecal/Soil Microbe Kits and Quick-RNA Plant Kit – optimising your workflow for sensitive applications like PCR and NGS.
Achieve reliable PCR results, free from inhibitors
PCR is a foundational technique in molecular biology, essential in applications from pathogen monitoring to crop trait analysis. Consistent, reliable amplification is crucial – and that starts with inhibitor-free nucleic acids.
Use Zymo Research’s OneStep PCR Inhibitor Removal Kit to ensure that PCR inhibitors are no longer limiting the quality or reproducibility of your results.
This article is based on an original post by Zymo Research. You can read the original here.
Featured supplier

Zymo Research
Zymo Research is a leader in molecular biology, offering a comprehensive range of products for DNA, RNA, and epigenetics research. Established in California in 1994, the company is renowned for its high-quality nucleic acid purification technologies, including kits and reagents for DNA and RNA clean-up, isolation, and sequencing. Zymo is also a pioneer in epigenetics, with products for DNA methylation analysis, chromatin analysis, and NGS library preparation. Each product is designed to be simple to use, reliable, and available at competitive prices, making them ideal for both academic and biopharmaceutical research.
Related products
| Cat-No. | Item | Size | Price (CHF) |
|---|---|---|---|
| D6030 | OneStep PCR Inhibitor Removal Kit | 50 preparations | 169.00 |
| R2001 | ZymoBIOMICS RNA Mini Kit | 50 preparations | 611.00 |
| R2002 | ZymoBIOMICS DNA/RNA Mini Kit | 50 preparations | 784.00 |
| D4300 | ZymoBIOMICS DNA Miniprep Kit (50 preps) Includes: [D4304] ZymoBIOMICS Miniprep Kit x 1, [S6012-50] ZR BashingBead Lysis Tubes 50 pack x 1 [D4300-1-40] ZymoBIOMICS Lysis Solution, 40 ml x 1 | 50 each | 394.00 |
| D4301 | ZymoBIOMICS DNA Microprep Kit (50 preps) Includes: [D4305] ZymoBIOMICS Microprep Kit x 1 [S6012-50] ZR BashingBead Lysis Tubes 50 pack x 1 [D4300-1-40] ZymoBIOMICS Lysis Solution, 40 ml x 1 | 50 each | 394.00 |
| D6010 | Quick DNA Fecal/Soil Microbe MiniPrep Kit | 50 preparations | 327.00 |
| D6012 | Quick-DNA Fecal/Soil Microbe MicroPrep Kit | 50 preparations | 327.00 |
| D6110 | Quick-DNA Fecal/Soil Microbe MidiPrep Kit | 25 preparations | 718.00 |
| D6020 | Quick-DNA Plant/Seed Miniprep Kit | 50 preparations | 327.00 |
| R2040 | Quick-RNA Soil/Fecal RNA MicroPrep Kit | 50 preparations | 496.00 |
| R2024 | Quick-RNA Plant MiniPrep Kit | 50 preparations | 424.00 |
| R2042 | Zymo Environ Water RNA Kit | 50 preparations | 574.00 |
Citations
- Schrader, C., Schielke, A., Ellerbroek, L., & Johne, R. (2012). PCR inhibitors - occurrence, properties and removal. Journal of applied microbiology, 113(5), 1014–1026.
- Sidstedt, M., Rådström, P., & Hedman, J. (2020). PCR inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Analytical and bioanalytical chemistry, 412(9), 2009–2023.
- Opel, K. L., Chung, D., & McCord, B. R. (2010). A study of PCR inhibition mechanisms using real time PCR. Journal of forensic sciences, 55(1), 25–33.
