MolBoolean is a novel fluorescent-based in situ protein proximity technology for the simultaneous detection of both interacting and non-interacting for two protein targets in cells and tissues. MolBoolean can extract more information from cell and tissue samples than previously possible. The method enables the simultaneous quantification of the relative amounts of individual as well as interacting proteins, thus making the cells' activity status and communication more visible.
MolBoolean™ Starter Kit
MolBoolean is available as a starter kit for researchers interested in evaluating the technology for their own applications. Starter kit highlights:
- Reagents for 40 cell and 20 tissue experiments
- Comprehensive, easy-to-follow protocol
- Free image analysis tool for result interpretation
- Optimized for quick integration into standard lab workflows
Interested? Contact our team
Advantages
- Complete spatial quantitative analysis of protein-protein interactions by simultaneous detection of free and interacting proteins.
- Accurate quantification by normalization of interaction data to total target protein levels.
- Biologically relevant data without the need for engineered protein expression.
- Able to detect and quantify low abundant proteins. Thanks to 1’000-fold increased fluorescence signal by Rolling Circle amplification.
- Universal kit that can be used with your choice of primary antibodies.
- Validated in both cells and tissue.
- Complementary analysis pipeline available in Open Source.
- Published and peer-reviewed technology.
- Adaptable to different research needs.
Product details
- Catalog Number: MolB00001
- Included in the Kit: Antibody proximity probes, oligonucleotides, enzymes, fluorescent detection reporters.
- Not included in the Kit: Primary antibodies.
- Target Species: Mouse, Rabbit.
- Kit Size: 4.8 ml, approximately 120 assays in cells (40 μl/assay) and 60 assays in tissue (80 μl/assay)
- Shipped on dry ice, storage at -20°C.
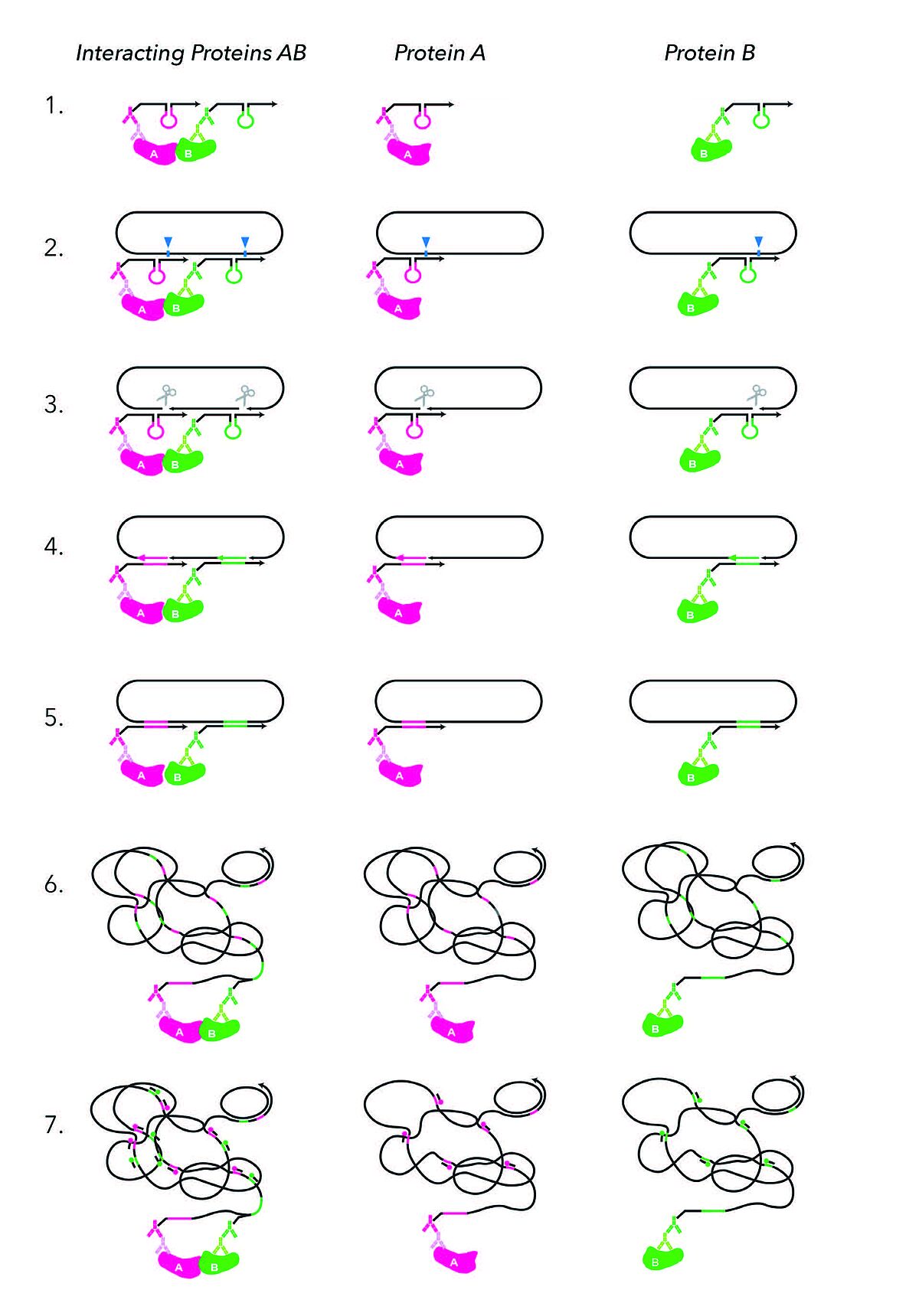
MolBoolean assay steps
The MolBoolean™ assay involves seven steps to detect whether the proteins of interest are interacting or present individually:
- Step: Proximity probe binding to primary antibodies.
- Step: Proximity probe arm hybridization to DNA circle oligo.
After binding their respective target proteins A and B, proximity probes A (black and magenta) and B (black and green) hybridize into the circle. Arrows signify oligonucleotide polarity. - Step: DNA nicking.
The circle gets enzymatically nicked (cyan arrowhead indicates nicking position). - Step: Tag oligo incorporation.
The circle gets invaded by reporter tags (tag A in magenta, tag B in green). - Step: DNA ligation.
Enzymatic ligation of the reporter tags to the circle follows. - Step: Rolling Circle Amplification.
Rolling circle amplification (RCA) creates long concatemeric products (RCPs). - Step: Detection.
RCPs are detected via fluorescently labeled tag-specific detection oligonucleotides.(1)
Application Examples
The MolBoolean technology was first published in Nature Communications by Raykova et al, (2022). This publication included several examples showcasing the sensitivity and specificity of the MolBoolean assay in both cells and tissue. Figure 2 and 3 showcase results obtained with MolBoolean on MCF7 cells and in human kidney, respectively.
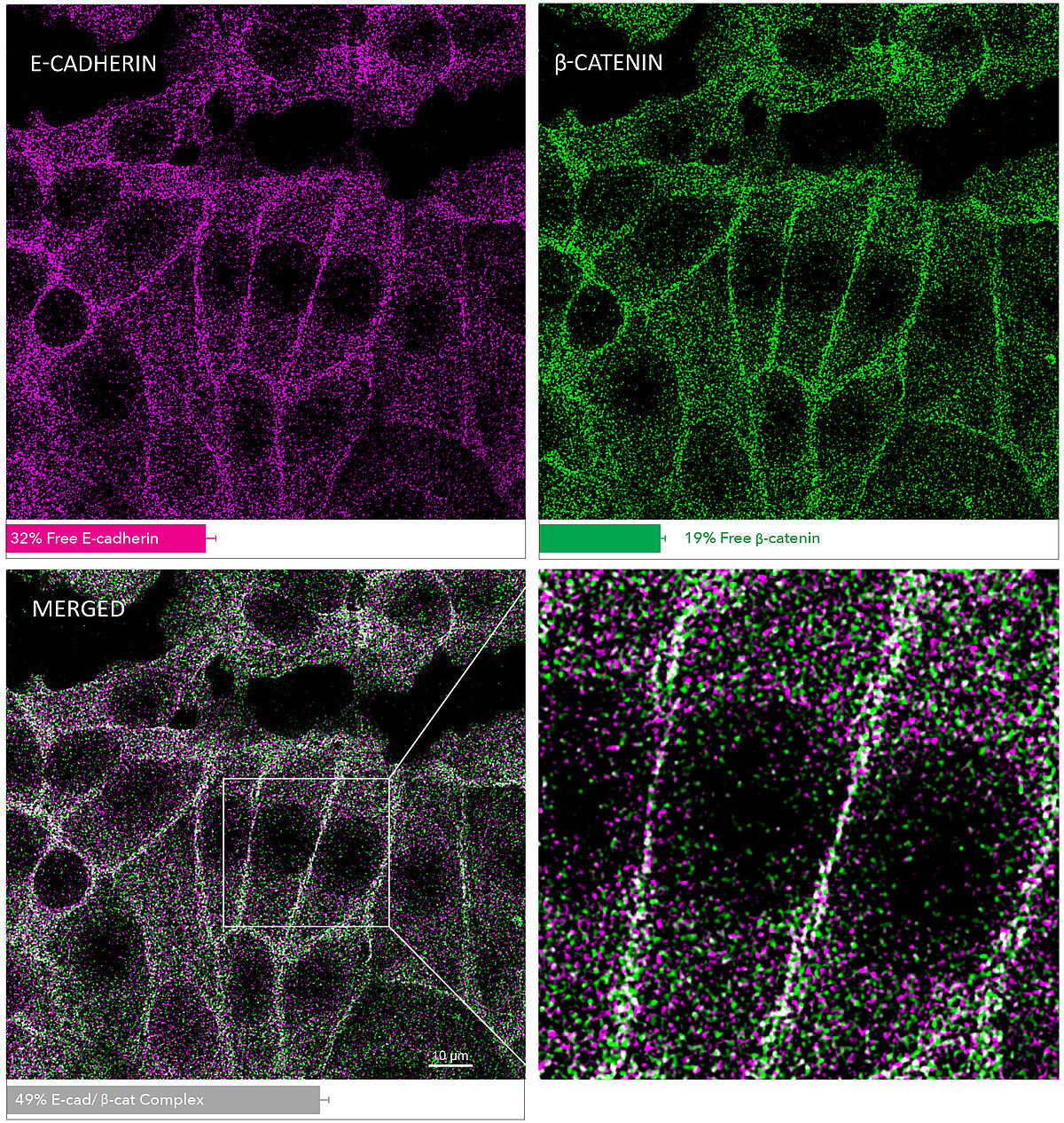
E-Cadherin/ β-Catenin MolBoolean Staining in MCF7 Cells
MolBoolean analysis of the interaction between E-cadherin (cat. AMAb90862, magenta) and β-catenin (cat. HPA029159, green) in MCF7 cells, showing the relative quantification of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels: 32% free E-cadherin (magenta), 19% free β-catenin (green), 49% E-cadherin/β-catenin complex (white). Data is normalized to total target protein levels (total RCPs).
ACE2/TMPRSS2 MolBoolean Staining in Human Kidney
MolBoolean analysis of the interaction between ACE2 (cat. AMAb91259, magenta) and TMPRSS2 (cat. HPA035787, green) in human kidney, showing the relative quantification of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels: 51% free ACE (magenta), 17% free TMPRSS2 (green), 32% ACE/TMPRSS2 complex (white). Data is normalized to total target protein levels (total RCPs).
Customize the secondary antibody species
You can now tailor the secondary antibody species in MolBoolean kits to better meet the specific needs of your customers. What this means for you:
- Personalized solutions: Offer kits that align precisely with your customers’ experimental requirements.
- Competitive advantage: Differentiate your offerings with unique, customizable features that help you stand out in the market.
Interested? Contact our team
References
- Raykova, D., Kermpatsou, D., Malmqvist, T. et al. A method for Boolean analysis of protein interactions at a molecular level. Nat Commun 13, 4755 (2022). doi.org/10.1038/s41467-022-32395-w