Scientists at Integrated DNA Technologies (IDT) published a comprehensive study into their CRISPR-Cas homology-directed repair (HDR) technology. Published in Nature Scientific Reports, Schubert et al. [1] describe how to maximise HDR efficiency using a set of design considerations which have been built into their novel bioinformatic tool for HDR donor template design.

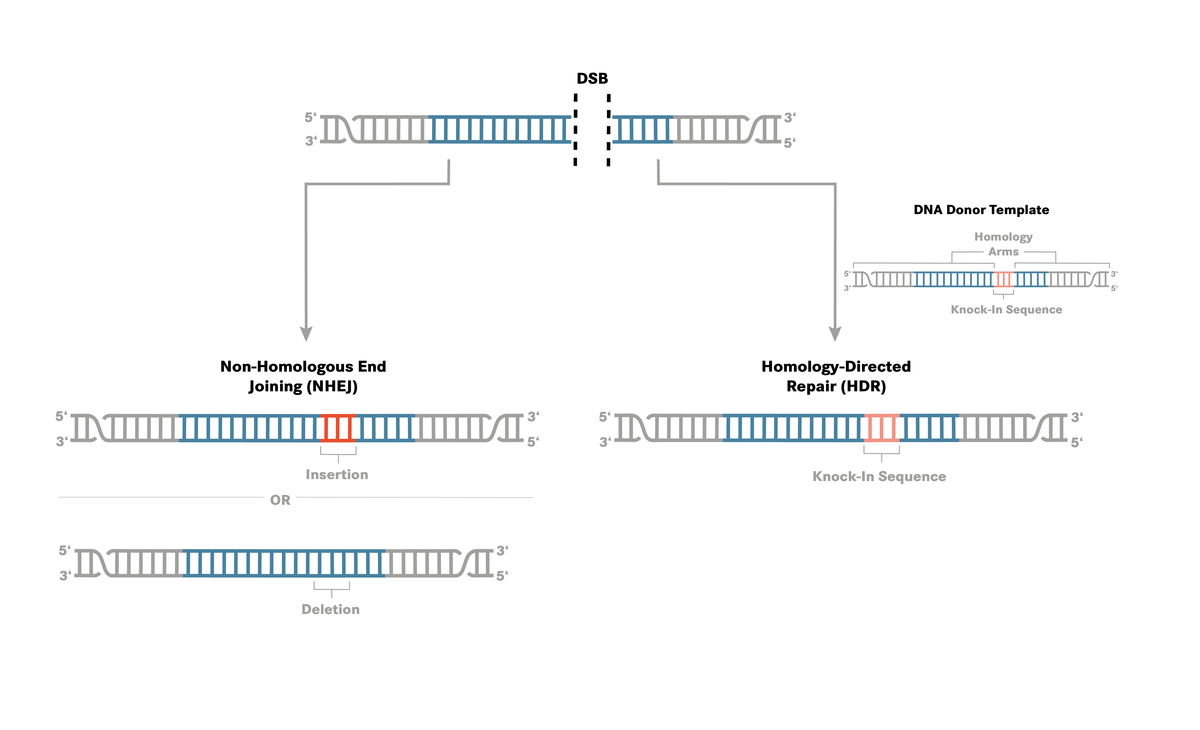
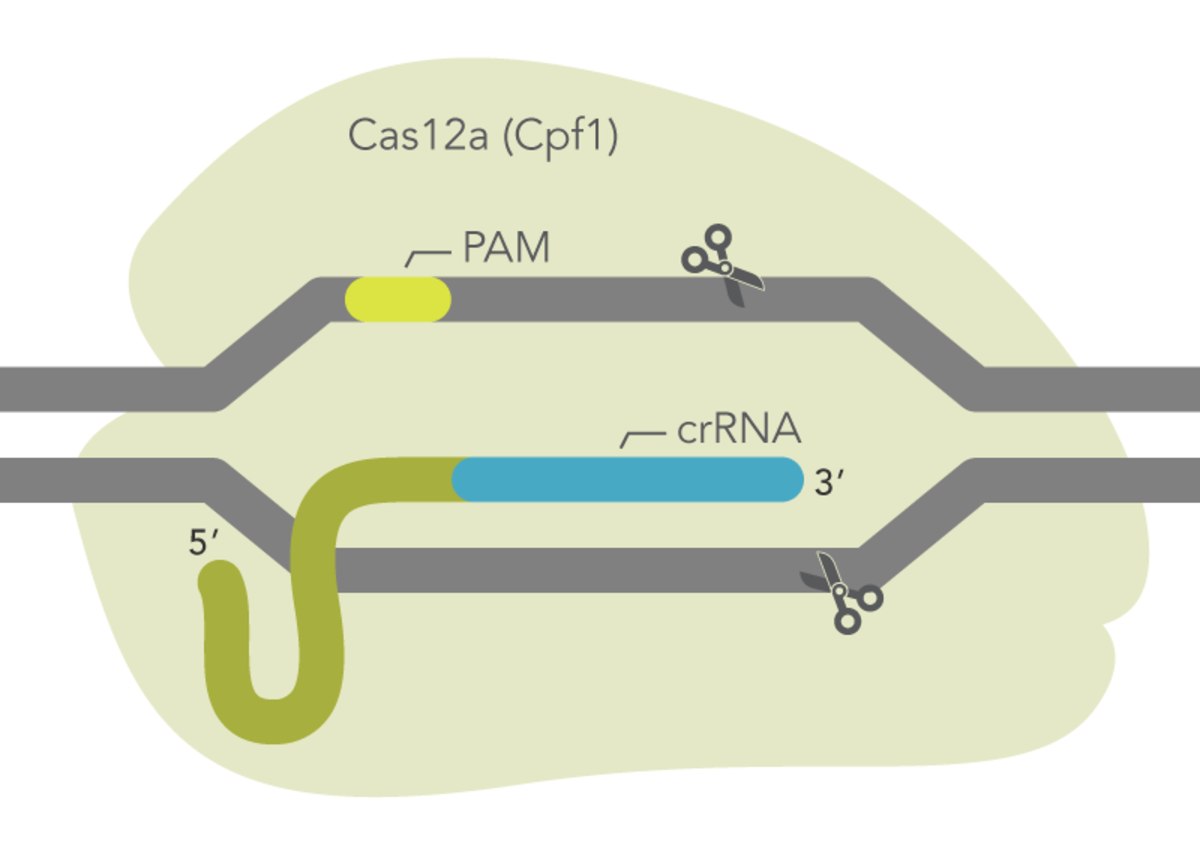
CRISPR (clustered regularly interspaced short palindromic repeats) technology is currently the most efficient method for site-directed genome editing, leading to permanent changes in genomic DNA. It is commonly used as a method for gene knockout by utilising a natural DNA repair mechanism known as non-homologous end joining (NHEJ). This 2021 publication concentrates on the competing HDR repair pathway which can be harnessed in the laboratory for CRISPR knock-in studies, protein tagging and mutation generation or repair. The HDR pathway can repair a double-stranded break (DSB) in DNA caused by Cas9 or Cas12a cleavage in an error free fashion. If a donor DNA template is added along with the ribonucleoprotein complex (Cas nuclease and guide RNA), insertion of the exogenous donor DNA is possible. The donor DNA can contain a gene of interest, a protein tag or a sequence designed to repair or introduce a mutation.
Homology-directed repair experiments can be challenging, the NHEJ pathway is the predominant mechanism for the repair of double-stranded DNA breaks, leading to poor rates of incorporation. Schubert et al. [1] investigated the optimal conditions to insert a mutation via a single-stranded oligodeoxnucleotide (ssODN) donor template. They looked at a number of variables to consider when designing a CRISPR-Cas homology-directed repair:
- Strand preference for the ssODN donor template – targeting strand (T strand; ssODN that is complementary to the CRISPR–Cas9 gRNA) or non-targeting strand (NT strand; ssODN that is non-complementary to the gRNA and contains the ‘NGG’ PAM sequence)
- Enzyme choice depending on the availability of a PAM site near to the desired mutation site.
- Use of blocking mutations to prevent re-cleavage by the CRISPR-Cas nuclease
- Ideal homology arm length
- Selection of gRNA(s) for each new HDR mutation location
To read their in-depth guidance on HDR design: Optimized design parameters for CRISPR Cas9 and Cas12a homology directed repair. Schubert et al. [1] 2021, Nature Scientific Reports
Alt-R CRISPR HSR Design Tool
The Alt-R CRISPR HSR Design Tool from IDT enables researchers to achieve industry leading HDR rates. Based on extensive research, this novel bioinformatic tool can be used to create and order HDR donor templates and associated Cas9 guide RNAs for genome editing of human, mouse, rat, zebrafish or C. elegans targets.
Alt-R™ products
The Alt-R™ range of products used in this publication are available through LubioScience.
IDT’s Alt-R™ CRISPR Cas9 and Cas12a systems are a complete solution for HDR based genome editing. Products include Cas nucleases, guide RNAs, enhancers and controls.
The Alt-R™ CRISPR-Cas9 System includes all the reagents needed for successful genome editing in your research applications, based on the natural S. pyogenes CRISPR-Cas9 system.
- Benefit from the latest improvements in on- and off-target design and chemical modifications, as well as easy ordering of custom or predesigned guide RNAs.
- Get optimal editing with high on-target potency and reduced off-target activity with Alt-R HiFi CRISPR-Cas9 nuclease
- Precisely control editing with efficient delivery of the RNP by lipofection or electroporation
- Help to protect your cells from toxicity or innate immune response activation
The Alt-R™ CRISPR-Cas12a System allows targeting of alternative sites that are not available to the CRISPR-Cas9 System and produces a staggered cut with a 5′ overhang.
- Enables genome editing in organisms with AT-rich genomes
- Allows interrogation of additional genomic regions compared to Cas9
- Requires simple complexing of crRNA with the Cas12a protein—no tracrRNA needed
- New - support for L.b. Cas12a crRNAs has been added to the ordering options
References
[1] - Schubert, M.S., Thommandru, B., Woodley, J. et al. Optimized design parameters for CRISPR Cas9 and Cas12a homology-directed repair. Sci Rep 11, 19482 (2021). doi.org/10.1038/s41598-021-98965-y