The challenge of read depths in NGS experiments
An important question in many next generation sequencing (NGS) experiments is how best to prepare samples so that the read depth is maximized for regions of interest (ROI) while the cost is kept low. Frequently, samples contain high concentrations of sequences that are not important to the conclusions of the sequencing project. For example, NGS projects that sequence DNA or RNA (i.e., RNA-seq) may be intended to determine a sequence of a virus, bacteria, or other pathogen found at low concentrations within a milieu consisting mostly of human sequences [1,2]. Another example is a situation in which ribosomal RNA makes up the majority of a eukaryotic sample; the sequences of such ribosomal RNA are often not of interest to the researchers [3–6]. Whole transcriptome sequencing in such samples would not be budget-friendly, it would slow down the rate of obtaining results, and it would decrease accuracy compared to sequencing a sample containing a higher percentage of the ROIs.
Therefore, finding and optimizing methods of preparing NGS samples to enrich the ROIs is an important area of research. Many such methods have been described; these methods often involve several steps of enzymatic interconversion of RNA and DNA, selectively using enzymes such as RNAse H to destroy RNA within RNA-DNA duplexes. Although many of these methods result in increases in read depth, they are often complex and may be applicable only to a subset of sample types. However, some of the newest approaches to improving read depth can greatly improve speed and simplicity. Several promising methods apply CRISPR-based technology, as described in more detail below.
A solution to the read depth problem - DASH
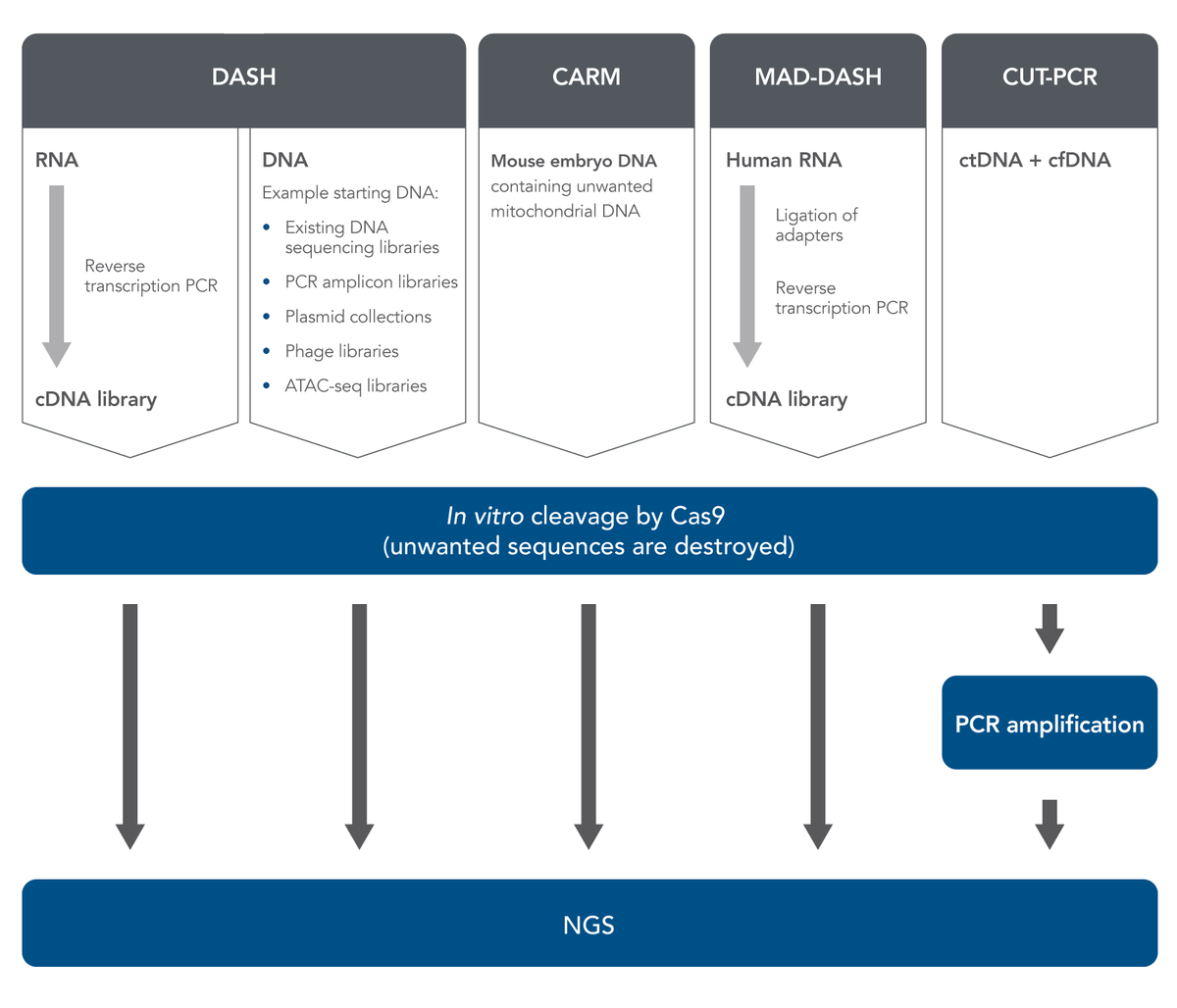
In 2016, Gu et al. published a method called Depletion of Abundant Sequences by Hybridization (DASH) [7]. (The word “hybridization” in this name refers to the hybridization of Cas9-bound gRNA with target DNA.) DASH is an in vitro method which can improve NGS sample prep, starting with either RNA or DNA (Figure 1). In the case of RNA, researchers start with biologically-derived mixed RNA specimens containing only small amounts of the sequences or regions of interest but large amounts of high-abundance, uninteresting sequences.
Reverse-transcription PCR is used to produce cDNA from the RNA. DASH then uses Cas9 nuclease (in vitro, not in cells or tissues), directed by gRNA, to cleave and destroy only the high-abundance, uninteresting cDNA sequences while leaving the rare cDNA ROIs intact. Therefore, these rare cDNA sequences become a higher percentage of the resulting sample. After this relative enrichment of rare sequences, next generation sequencing can quickly and less expensively detect these sequences.
Gu et al. showed that this method could remove 12S and 16S mitochondrial rRNA in samples obtained from HeLa cells. They then demonstrated successful removal of large amounts of 12S and 16S mitochondrial rRNA from patients’ cerebrospinal fluid in order to enrich and detect very low concentrations of pathogenic amoebal, fungal, or tapeworm RNA sequences. The coverage depth for each kind of pathogenic sequence was increased several fold by this process.
Gu et al. pointed out the potential superiority of DASH over non-CRISPR-based methods of RNA sample preparation for targeted sequencing based on the fact that, when starting with low total amounts of RNA, the reverse transcription PCR to produce cDNA amplifies the total sample amount. This allows researchers to start with any very low amount of sample, unlike non-CRISPR-based methods that require a minimum of 10 ng of RNA as starting material.
After performing DASH, Gu et al. performed metagenomic deep sequencing. They showed that DASH had significant performance, programmability, and cost benefits compared to non-CRISPR based methods of sample enrichment methods.
Applications of DASH
Gu et al. also showed that the DASH method can be used to aid in sample preparation when starting with genomic DNA samples, not just RNA-derived cDNA [7]. They used a human genomic DNA sample which they had mixed with small amounts of mutant KRAS G12D DNA. In this case, the DASH approach depleted the wild-type DNA, enriching the mutant DNA, thus allowing for downstream detection by digital droplet PCR (ddPCR). The DASH method, therefore, was shown to be useful not only with cDNA derived from RNA, but was also able to eliminate large amounts of genomic DNA from a sample containing low levels of mutant DNA.
Prezza et al. used the DASH method in a study of bacterial RNA [8]. In agreement with Gu’s findings, the DASH method was useful when other methods of depleting ribosomal RNA could not be used; in particular, DASH was useful even when only sub-nanogram levels of cDNA were available. Furthermore, the DASH method was specifically useful for studies of bacterial RNA transcripts, as the method did not depend on polyadenylation, unlike some others.
In a study of mammalian preimplantation chromatin, Wu et al. used a method much like DASH to deplete mitochondrial DNA for specific analysis of accessible chromatin. They used a modified version of the Assay for Transposase-Accessible Chromatin with high throughput sequencing (ATAC-seq) and named it CARM (CRISPR/Cas9-Assisted Removal of Mitochondrial DNA) [9]. They then validated CARM and showed that it exhibits high sensitivity. Montefiori et al. used a similar DASH-like ATAC-seq approach to reduce mitochondrial DNA reads in samples from lymphoblastoid cell lines [10]. Their CRISPR-based, DASH-like method outperformed a method based on detergent removal, and it decreased the amount of sequencing needed to get the highest quality data.
Hardigan et al. have pointed out that during preparation of microRNA (miRNA) samples for NGS, a specific problem frequently occurs; namely, formation of adapter dimer ligation products, which are only about 20–30 bases shorter than the miRNAs to be sequenced. These adapter dimer ligation products can dilute the ROIs and decrease NGS sensitivity. To address this known problem, Hardigan et al. adapted the DASH method specifically for depletion of these unwanted small RNA (smRNA) sequences [11]. By removing adapter dimer ligation products, they improved quantitation of miRNA found at low levels in plasma or tissue samples. They termed their method miRNA and Adapter Dimer–DASH (MAD-DASH).
Another method similar to DASH, CRISPR-mediated, Ultrasensitive detection of Target DNA-PCR (CUT-PCR), was developed by Lee et al. to clean up human plasma-derived DNA for liquid biopsy cancer detection [12]. These researchers reasoned that since circulating tumor DNA (ctDNA) can be found in human plasma, and is usually at much lower concentrations than normal circulating cell-free DNA (cfDNA), various Cas nucleases including S.p. Cas9 or F.n. Cas12a could be used to cleave and destroy cfDNA, leaving the ctDNA untouched. They focused on cleaving sequences in which the protospacer-adjacent motif (PAM) site was mutated in the ctDNA. After cleaving DNA samples with Cas nucleases, they amplified the remaining sequences by PCR to enrich the ctDNA sequences for targeted deep sequencing. Therefore, the CUT-PCR method can successfully be used to eliminate large amounts of naturally-occurring normal DNA in plasma in order to enrich ctDNA. This may increase sensitivity and lower the limit of detection in liquid biopsies, which would enable earlier cancer screening.
Summary
The CRISPR-associated (Cas) enzymes can be considered very highly tunable and flexible restriction enzymes—they can cut any desired target sequence. Thus, researchers can use them to deplete unwanted sequences, resulting in enrichment of rare sequences. The above methods of using CRISPR to deplete unwanted DNA sequences for downstream targeted NGS analysis demonstrate the great flexibility of CRISPR as a tool for research and medicine. IDT can supply the guide RNA and the Cas enzymes needed for further research using CRISPR either for depletion of unwanted sequences or for many other applications.
References
- Hasan MR, Rawat A, et al. (2016) Depletion of human DNA in spiked clinical specimens for improvement of sensitivity of pathogen detection by next-generation sequencing. J Clin Microbiol 54(4):919–927.
- Nelson MT, Pope CE, et al. (2019) Human and extracellular DNA depletion for metagenomic analysis of complex clinical infection samples yields optimized viable microbiome profiles. Cell Rep 26(8):2227–2240.e2225.
- Fauver JR, Akter S, et al. (2019) A reverse-transcription/RNase H based protocol for depletion of mosquito ribosomal RNA facilitates viral intrahost evolution analysis, transcriptomics and pathogen discovery. Virology 528:181–197.
- Adiconis X, Borges-Rivera D, et al. (2013) Comparative analysis of RNA sequencing methods for degraded or low-input samples. Nat Methods 10(7):623–629.
- Petrova OE, Garcia-Alcalde F, et al. (2017) Comparative evaluation of rRNA depletion procedures for the improved analysis of bacterial biofilm and mixed pathogen culture transcriptomes. Sci Rep 7(1):41114.
- Zhao S, Zhang Y, et al. (2018) Evaluation of two main RNA-seq approaches for gene quantification in clinical RNA sequencing: polyA+ selection versus rRNA depletion. Sci Rep 8(1).
- Gu W, Crawford ED, et al. (2016) Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol 17(1).
- Prezza G, Heckel T, et al. (2020) Improved bacterial RNA-seq by Cas9-based depletion of ribosomal RNA reads. RNA doi: 10.1261/rna.075945.075120.
- Wu J, Huang B, et al. (2016) The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534(7609):652–657.
- Montefiori L, Hernandez L, et al. (2017) Reducing mitochondrial reads in ATAC-seq using CRISPR/Cas9. Sci Rep 7(1):2451.
- Hardigan AA, Roberts BS, et al. (2019) CRISPR/Cas9-targeted removal of unwanted sequences from small-RNA sequencing libraries. Nucleic Acids Res 47(14):e84–e84.
- Lee SH, Yu J, et al. (2017) CUT-PCR: CRISPR-mediated, ultrasensitive detection of target DNA using PCR. Oncogene 36(49):6823–6829.