Antimicrobial resistance (AMR) is a rising worldwide health threat that can render antibiotics ineffective. This phenomenon limits treatment options clinicians can offer patients to clear up bacterial infections, threatening seven decades of progress treating devastating infectious diseases and pressuring researchers to figure out how to stifle the occurrence of AMR and develop new treatment modalities in preparation of antibiotics becoming obsolete.
The problem
Antimicrobial resistance is often associated with antibiotic treatment putting pressure on microbes to adapt (mutate) to survive. We now know that individuals can acquire AMR genes regularly found in their destinations and bring them back home. Because AMR bacteria can pass their AMR genes to other bacteria in the human gut, introducing new AMR genes into regions can lead to a higher prevalence of antibiotic resistance among the community through human contact and shared environments.
Returning travelers are rarely tested for AMR genes or resistant bacteria unless they are sick, so the magnitude of AMR gene acquisition risk from international travel remains unknown, but to fully address a problem, we need to understand the scale of the issue as well as key characteristics and drivers.
An AMR gene’s ability to spread via international travel depends on:
- the AMR gene’s prevalence in the endemic region
- the specific bacteria harboring the AMR gene
- and the other genetic elements colocalized with the gene
The solution
To gain insight to how international travel influences the gut microbiome and AMR gene acquisition, D’Souza et al., evaluated at the abundance, diversity, function, resistome architecture, and context of AMR genes in international travelers.
The approach
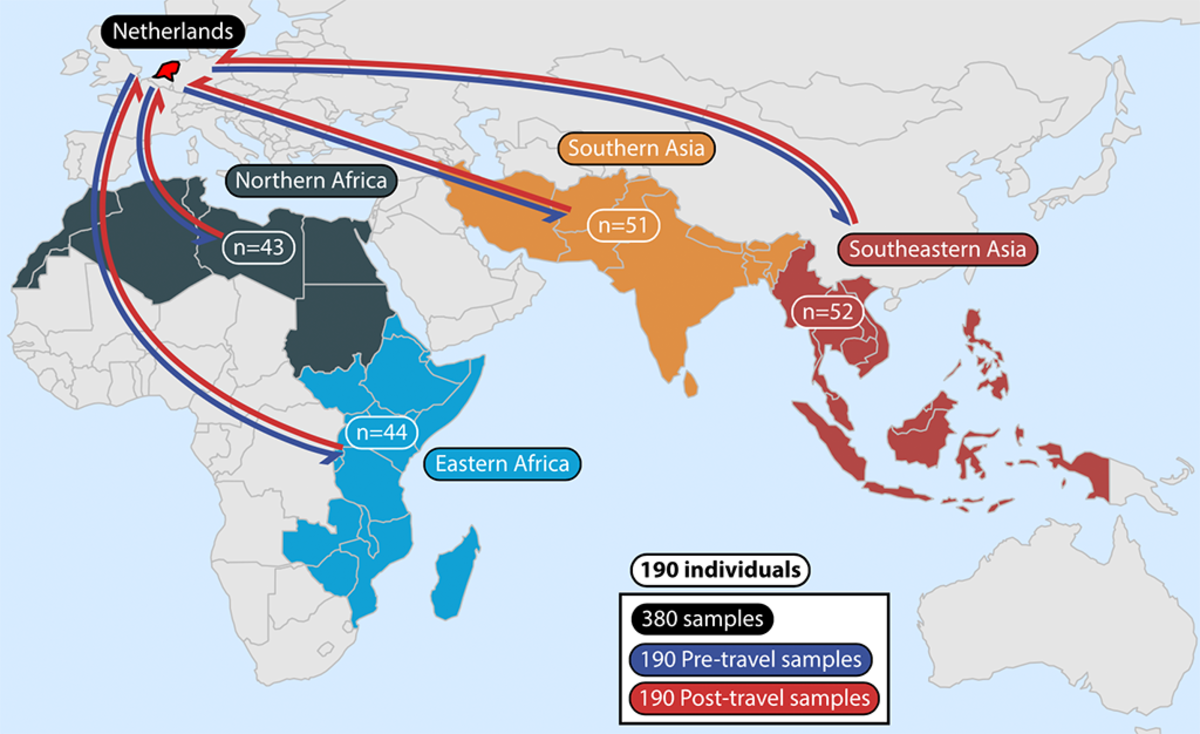
Using whole metagenomic shotgun sequencing and functional metagenomics the researchers studied multiple aspects of the gut microbiome in 190 Dutch individuals from fecal samples before and after travel to diverse locations around the world, including:
- Northern Africa (n = 43)
- Eastern Africa (n = 44)
- Southern Asia (n = 51)
- Southeastern Asia (n = 52)
Why Functional Genomics?
Using functional metagenomic selection offers higher sensitivity to detect AMR genes that may be missed by conventional metagenomics due to underrepresented in established AMR databases. This is reflected in D’Souza’s et al. results where 51 of the 121 (42.1%) AMR genes detected and compared in the analysis were found via functional selections.
The findings
The researchers found that international travel significantly altered the gut resistome and showed that the traveler's resistomes and AMR gene acquisitions are destination specific.
Following international travel,
- AMR gene abundance and α-diversity increases, but β-diversity decreased
- AMR gene abundance increased and acquisitions fell within several AMR gene families and resistance mechanisms
- Those that went to Southeast Asia had the highest AMR gene acquisition and had high mobile genetic element burden
In terms of gene acquisition, the researchers found that acquisition events mainly consisted of AMR genes with efflux, inactivation, and target replacement resistance mechanisms, and AMR gene acquisition colocalizes with mobile genetic elements. Some of the high-risk, mobile genetic element-associated AMR genes include qnr fluoroquinolone resistance genes, blaCTX-M family extended-spectrum β-lactamases, and plasmid-borne mcr-1 colistin resistance gene.
Here are some highlights of what the researchers found
- Travel increased the abundance and α-diversity of AMR genes in the traveler’s gut resistome, with 56 unique AMR genes showing significant acquisition after international travel and 4 AMR genes lost after travel
- Tetracycline inactivation AMR genes increased in abundance after travel, but the acquisition was only significant for one of two tetX variants, which was only detected by functional selections
- Plasmid-borne colistin resistance gene mcr-1 was only acquired by those that traveled to Southeastern Asia
- Individuals returning to The Netherlands from the same destination country were more likely to have similar resistome features
The impact
The results show that travel shapes the human gut resistome and can lead to AMR gene acquisition against diverse antibiotics—threatening public health by the global spread of locally endemic AMR genes.
This study also reflects the importance of being aware that there are large geographic differences in the prevalence and type of resistant bacteria and their AMR genes that can be acquired by visiting travelers. Generally, low- and middle-income countries have higher endemic AMR than high-income countries due to antibiotic overuse.
While this study studied AMR genes that travelers acquire, it is absolutely feasible that travelers could pass their AMR genes to locals at their destinations. To further connect the dots between AMR gene transmission networks, further investigations are needed.
Read the full article
D’Souza, Alaric W., et al. "Destination shapes antibiotic resistance gene acquisitions, abundance increases, and diversity changes in Dutch travelers." Genome medicine 13.1 (2021): 1-21. https://doi.org/10.1186/s13073-021-00893-z