The Problem
SARS-CoV-2 (COVID-19) has infected 64 million people worldwide, leading to almost 1.5 million deaths, according to John Hopkins Research Center. Understanding the mechanism of infection is critical to designing effective drugs and vaccines. A key question is: What human genes are required for SARS-Cov-2 infection? Currently, only a few genes, such as ACE2 and cathepsin L, are known to be essential host genes for SARS-CoV-2 infection, which is a barrier to developing effective therapeutic strategies to fight this pandemic.
What is SARS-CoV-2?
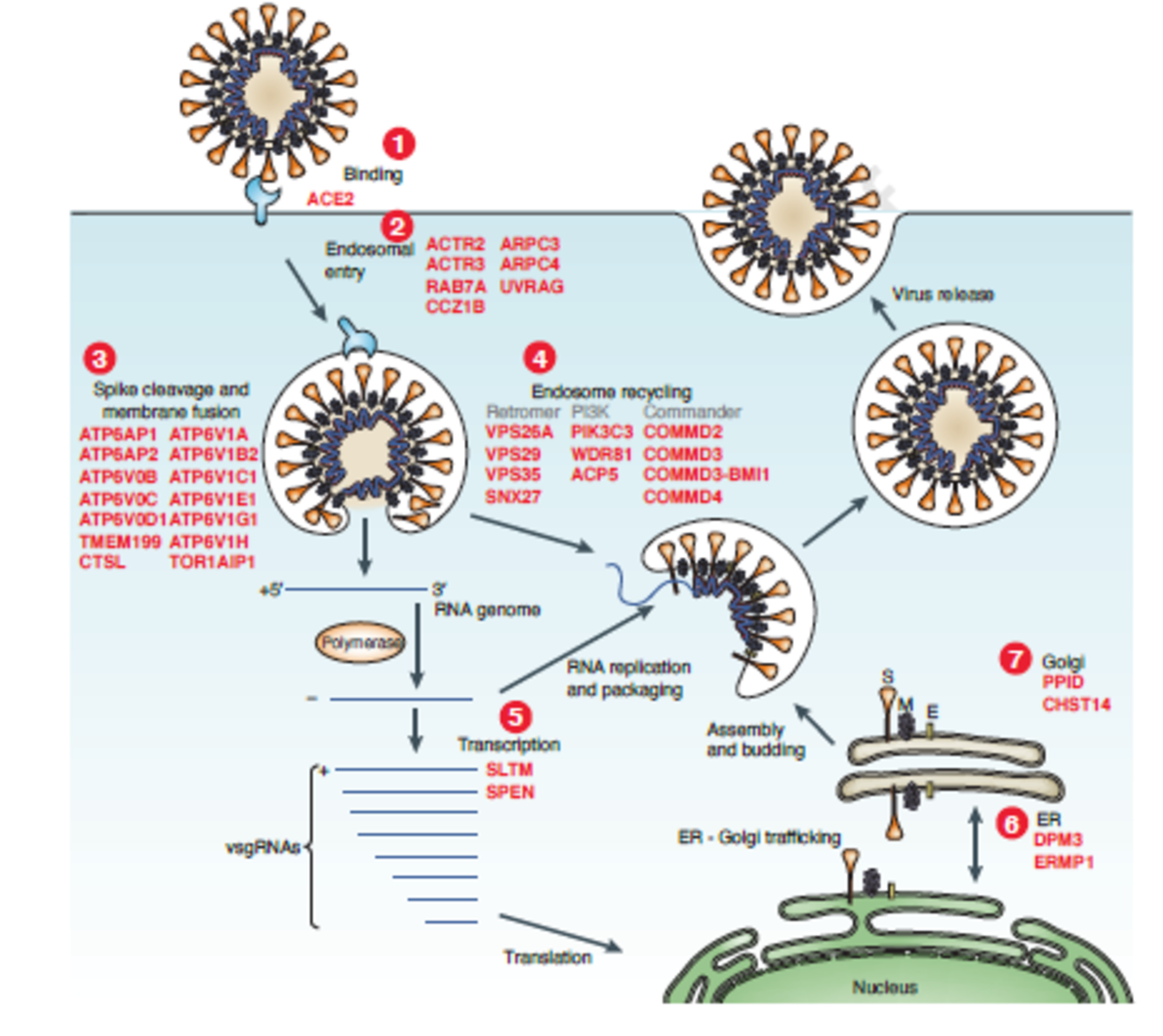
SARS-CoV-2 is an enveloped positive-sense RNA virus that is coated by Spike protein trimers that bind host angiotensin converting-enzyme2 (ACE2). Spike protein is proteolytically cleaved (catalyzed by many host proteases) allowing for viral-host membrane fusion and the release of viral RNA into the host cytoplasm, where it hijacks host machinery to generate more viral particles.
The approach and findings
Daniloski, et al. used a genome-scale CRISPR loss-of-function screen to identify targets among host genes that are required for SARS-CoV-2 infection1.
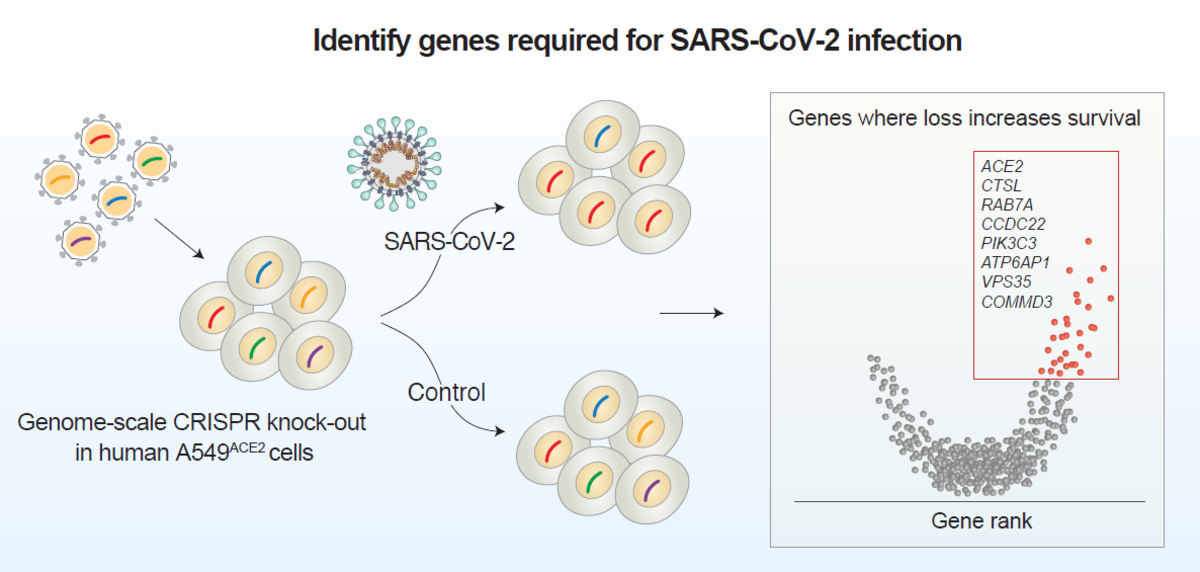
Identify genes required for SARS-CoV-2 infection
The researchers targeted 19,050 human genes using the GeCKOv2 CRISPR-Cas9 library, which contains 122,411 single-guide RNAs. They transduced a human epithelia carcinoma cell line that constitutively expresses ACE2 with a lentiviral vector containing Cas9, guide RNAs, and puromycin resistance gene. Cells were infected with either a high or low dose of SARS-CoV-2 and genomic DNA was extracted for amplicon sequencing to quantify guide RNA abundance. Without CRISPR-mediated gene perturbation, SARS-CoV-2 rapidly killed cells.
Using robust-rank aggregation (RRA) on the guide relative enrichments, the researchers computed gene-level scores to identify genes where loss-of-function led to enrichment within the pool. The top 50 most enriched genes were identified, and the researchers found 27 genes that were shared between the low and high viral dose conditions, suggesting that several host genes function independently of viral dose.
The 50 top-ranked genes clustered into distinct pathways, including:
- Initial attachment and endocytosis
- Spike protein cleavage and viral membrane fusion
- Endosome function, regulation, and recycling
- ER-Golgi trafficking
- Transcriptional modulation
Additionally, several of the highly-ranked genes from the loss-of-function screen were highly enriched in prior screens for Zika virus and pandemic H1N1 avian influenza. Gene ontology categories of enriched genes for SARS-CoV-2 was more similar to Zika virus than to H1N1.
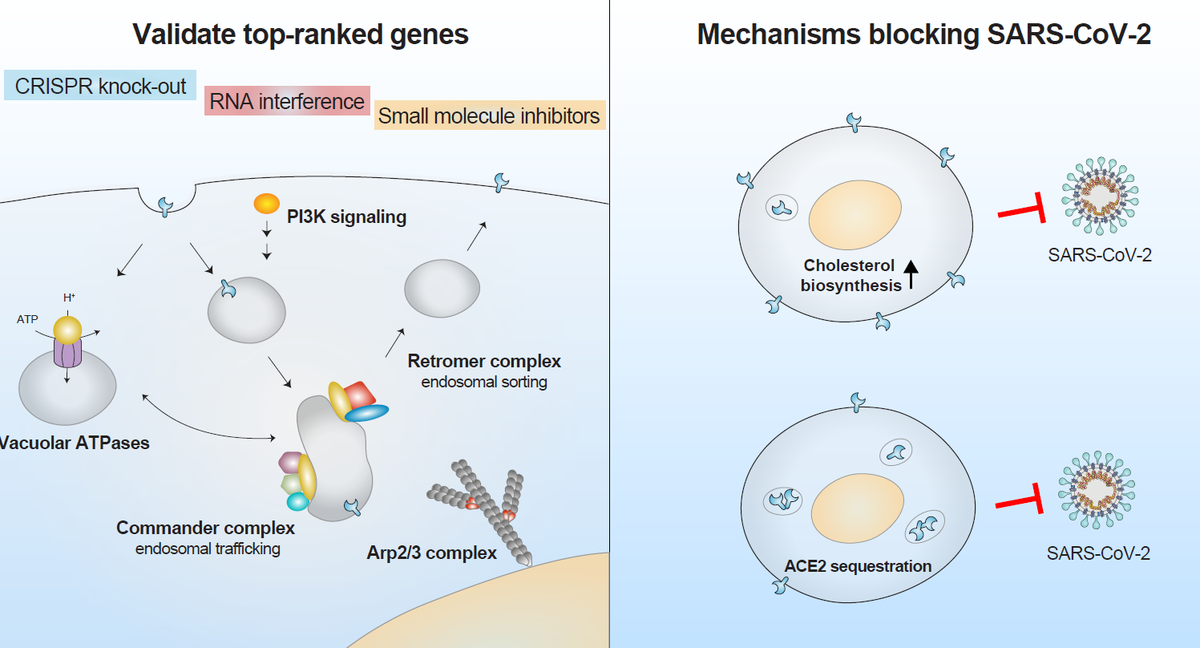
Validate top-ranked genes
Top-ranked genes were validated using several orthogonal methods including CRISPR knock-out, RNA interference, and small-molecule inhibitors. The researchers independently targeted the 30 top-ranked genes using CRISPR Cas9-mediated loss-of-function and assessed protein expression in cell lines infected with SARS-CoV-2. CRISPR Cas9-mediated knockout lowered the percentage of infected cells compared with cell lines with non-targeting guide RNAs, with up to a 10-fold reduction in SARS-CoV-2 infection.
The greatest protection was observed in cells lacking vesicular trafficking genes (RAB7A, CCDC22, VPS35) and in cells lacking ACE2 receptor and protease cathepsin L. Using the same approach in liver cells produced a similar reduction in infection. The researchers further validated this effect using siRNA knockdown for numerous genes.
Of the highly-ranked genes, 69 were considered druggable and 9 were selected for validation with small molecule inhibitors, as each were a primary or secondary target for at least one of the 26 inhibitors used in the study. Seven of the inhibitors tested reduced viral load more than 100-fold and two (autophinib and ALLN) reduced viral load more than 1000-fold. Four of the top performing molecules target the PIK3C3 gene, making it, or the endosomal entry pathway, a promising drug target.
Explore mechanisms blocking SARS-CoV-2
Using single-cell RNA-sequencing, the authors identified similar transcriptional changes in cholesterol biosynthesis upon the loss of top-ranked genes. Lacking one of six highly-ranked endosomal entry pathway genes—ATP6AP1, ATP6V1A, CCDC22, NPC1, PIK3C3 and RAB7A—upregulate pathways affecting lipid and cholesterol homeostasis, and can increase cholesterol by 10-50% depending on the specific gene. Interestingly, the researchers found that pre-treatment with amlodipine—a calcium-channel antagonist that upregulates cholesterol levels—reduced SARS-CoV-2 viral infection, which aligns with recent clinical studies suggesting patients taking amlodipine have a lower COVID-19 fatality rate2-3.
The impact
Daniloski et al. offer a genome-scale, quantitative resource covering the impact of the loss of each host gene on response to viral infection with COVID-19. This resource could potentially help research institutions, governmental organizations and pharmaceutical companies identify and develop antiviral drugs and vaccines.
Read the full publication: https://doi.org/10.1016/j.cell.2020.10.030
References
- Daniloski, Zharko, et al. "Identification of required host factors for SARS-CoV-2 infection in human cells." Cell (2020).
- Solaimanzadeh, Isaac. "Nifedipine and Amlodipine Are Associated With Improved Mortality and Decreased Risk for Intubation and Mechanical Ventilation in Elderly Patients Hospitalized for COVID-19." Cureus 12.5 (2020).
- Zhang, Leike, et al. "Calcium channel blocker amlodipine besylate is associated with reduced case fatality rate of COVID-19 patients with hypertension." medRxiv (2020).