Introduction
First introduced by IDT in 2012, double-stranded DNA (dsDNA) fragments provide a faster and more costeffective alternative to traditional gene synthesis, allowing researchers to drive down the cost and time of cloning experiments. Today, there are many commercially available dsDNA fragments on the market with varying degrees of fidelity. Unlike traditional gene synthesis products, dsDNA fragments are not clonal and are typically delivered as a mixed population of sequences, including a percentage of sequences containing mutations and truncations.
To meet the growing demands of researchers, IDT offers three different types of dsDNA fragments:*
• gBlocks™ Gene Fragments between 125–3000 bp in length
• eBlocks™ Gene Fragments between 300–900 bp, best suited for high-throughput screening and antibody discovery applications
• gBlocks HiFi Gene Fragments between 1000–3000 bp, optimized for large gene construction This paper will focus on the efficiency of IDT dsDNA fragments—gBlocks and gBlocks HiFi—as compared to dsDNA fragments from a few other synthesis providers.
Errors lead to experimental inefficiency
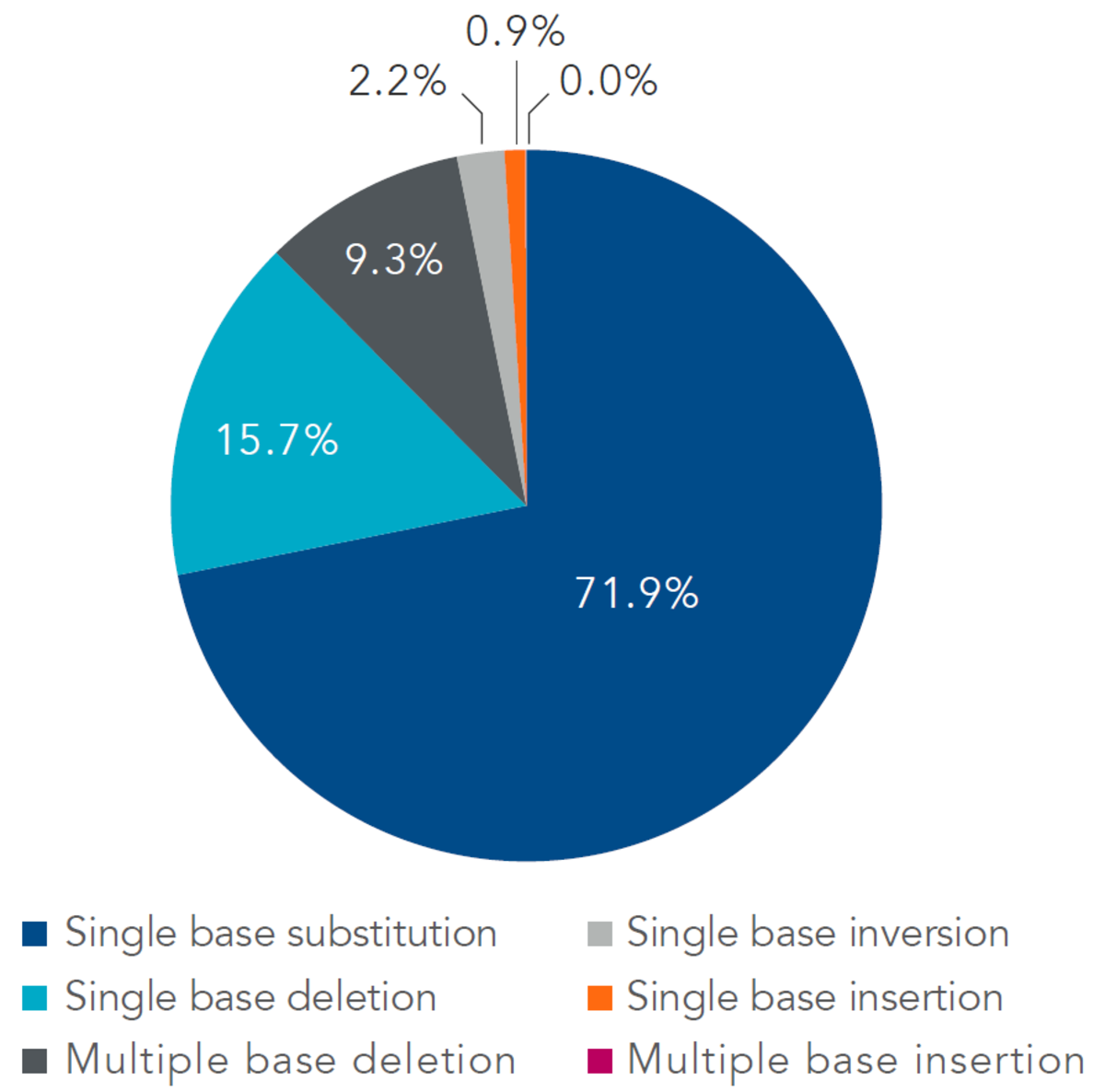
A common research application of these dsDNA fragments is cloning into a bacterial vector for the purpose of expressing a protein. Mutated species in a mixed population of dsDNA fragments can have negative effects on the results of an experiment. Mutations can lead to codon changes, frameshifts, and even complete expression failure. While all types of mutations are possible, single base substitutions, or deletions, randomly distributed through the population of dsDNA fragments tend to be the most common DNA errors observed from commercially available fragments (Figure 1). These errors are often expressed as the ratio of incorrect to correct bases and may be introduced from the oligonucleotides used to assemble the fragments or by the assembly process itself. Likewise, additional PCR amplification of the fragment or the cloning process itself may introduce errors.
Figure 1. Single base substitutions account for the largest percentage of observed errors. Distribution of gBlocks Gene Fragment errors was determined based on next generation sequencing (NGS) analysis of 96 gBlocks Gene Fragments ranging from 149 to 840 bps in length. Greater than 90% of the observed errors are single point mutations, with the majority of these mutations in the form of substitutions.
To compensate for these DNA sequence errors, researchers will often pick and sequence multiple clones or colonies of bacteria to ensure that their desired sequence can be found. Since the errors tend to be random in nature, the higher the inherent error rate, the more colonies a researcher needs to select to increase the probability of finding the desired sequence. The random nature of the errors also means that the length of the insert is a factor, as longer insert sizes increase the probability of a random error being present. Therefore, as the length of insert increases, so too must the number of colonies researchers screen. Increased screening time and costs add to the total time and cost of an experiment. In this paper, we will demonstrate how the proprietary synthesis and sequence error correction processes of IDT result in lower error rates and why these processes lead to a reduction in screening effort to find a correct clone when compared to alternative DNA synthesis providers in the market.
The IDT error correction method
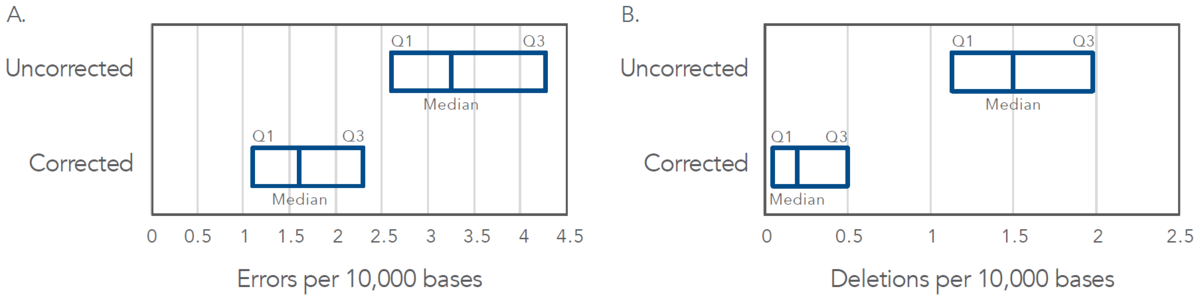
The majority of dsDNA fragments on the market are assembled using single-stranded oligonucleotides (oligos). Any mutation in the oligo then has the possibility of still being present in the assembled double-stranded construct. As the premier provider in oligo synthesis for over 30 years, IDT has robust synthesis processes which produce industryleading oligos. In addition to starting with high-fidelity oligos in our dsDNA assembly processes, IDT has developed a proprietary error correction method that is used in the manufacturing of all our double-stranded oligos (eBlocks, gBlocks, and gBlocks HiFi Gene Fragments). This error correction method has been shown to virtually eliminate deletions and greatly reduce errors of all types (Figure 2).
Figure 2. gBlocks Gene Fragments demonstrate lower total error rates and reduced deletion rates. Based on an NGS analysis using Illumina™ MiSeq™ sequencing of 218 gBlocks Gene Fragments ranging from 144 to 837 bases in length, the IDT proprietary error correction methods show a greater than 50% reduction in median error rate (A) and a greater than 85% reduction in deletion rate (B) Q1, 25th percentile; Q3, 75th percentile.
Quality assessment method
To explore the benefit this correction has on cloning performance, IDT ordered eight fragments with lengths ranging from 300 bp to 1750 bp from other leading DNA fragment suppliers. Then, IDT manufactured the same sequences as gBlocks Gene Fragments for sequences under 1 kb, and as gBlocks HiFi Gene Fragments for sequences over 1 kb using our standard manufacturing pipeline. To highlight the effect of sequence composition on error rates and cloning efficiency, sequences were designed at each length range with both balanced GC (median GC% of 50.2%) and lower GC (median GC% of 40.2%).
All fragments were diluted to a fixed concentration using a TE pH 8.0 buffer. A length verification was performed using capillary electrophoresis on an Agilent Fragment Analyzer (FA). Following length verification, the sample was prepped for sequencing using a Nextera™ XT DNA Library Preparation Kit (Illumina™) and run on an Illumina™ MiSeq™ instrument using a 2 x 150 read length to determine error rate. Error rate was calculated by dividing the total number of errors by the total number of sequenced bases.
All sequences were then cloned into a pUC based vector using a seamless cloning method, transformed, and then grown overnight on agar plates. Twenty-four colonies of each sequence were selected and prepared using a Nextera™ XT DNA Library Preparation Kit and run on an Illumina™ MiSeq™ instrument to determine their sequence; poor-quality reads were discarded.
Quality assessment of DNA fragments
To assess the overall quality of the fragments, we used two metrics: capillary electrophoresis (CE) to determine length and sequence verification by NGS. We started our analysis by reviewing the data using the Agilent FA capillary electrophoresis system. As shown in the electronic gel images created with the FA (Figure 3), we observed strong bands corresponding to the correct lengths on the ladder, indicating that all fragments contain a full-length product of the correct length. One fragment—a 1750 bp fragment from Supplier A—also contained a band corresponding to a length of approximately 500 bps. This indicates the presence of a sub-population containing a smaller fragment. In many applications, these truncated fragments can preferentially clone, resulting in incorrect colonies.
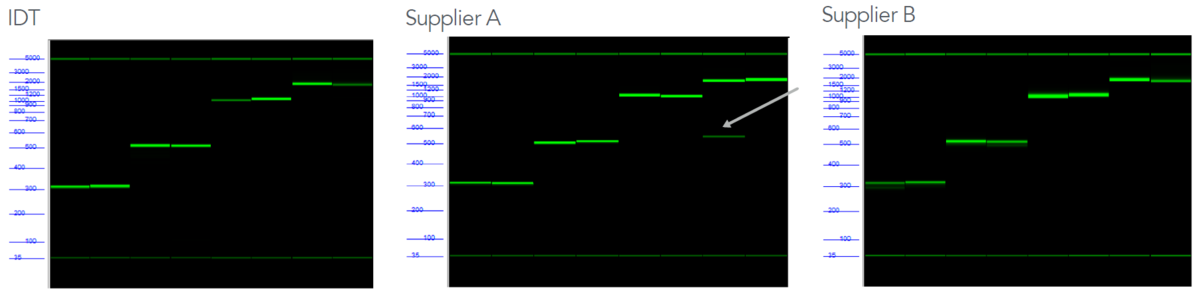
Figure 3. Another supplier’s dsDNA fragments show the presence of truncated product in gel image output. Outputs from the Fragment Analyzer show strong bands from all suppliers, corresponding to the correct length on the ladder. However, in the sample from Supplier A, a second smaller band is seen, indicating the presence of a truncated product.
Sequence composition of dsDNA fragments
After assessing the lengths of the individual fragments, the next step was to assess the sequence composition. Double-stranded fragments typically contain a mixed population. If the mixed population includes a significant truncated product, it can be detected by CE or by running on a gel. Commonly, fragments contain randomly distributed point mutations with any given mutation appearing only on a few molecules. To assess these mutations, we sequenced each fragment using an Illumina™ MiSeq™ instrument (Figure 4).
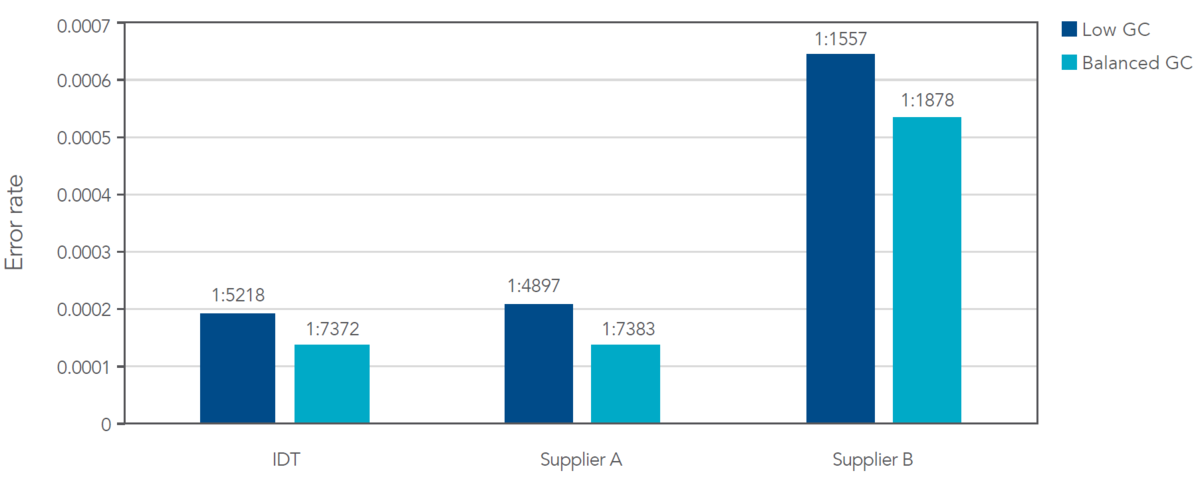
Figure 4. Lowest error rates compared to two other suppliers. Based on the NGS analysis of dsDNA fragments between 300 and 1750 bps in length, IDT fragments showed the lowest error rate in both balanced and low GC samples. In addition, a low GC content can increase error rates. N = 4 per supplier per condition.
The effect of lower GC content can be observed in the sequences from all three suppliers. The sequences containing lower GC content contained higher error rates when compared to the sequences containing balanced GC. In both scenarios, IDT fragments displayed the lowest average error rate. The average error rate for IDT fragments was ~1 in 6300, a 73% lower error rate than obtained with fragments from Supplier B.
Sequence errors affect experimental outcomes
To explore how such error rates affect experimental outcomes in a common application, each fragment was cloned into a pUC-based vector, and 24 colonies of each sequence were picked and sequenced to determine the number of clones that contained the correct desired sequence (Figure 5).
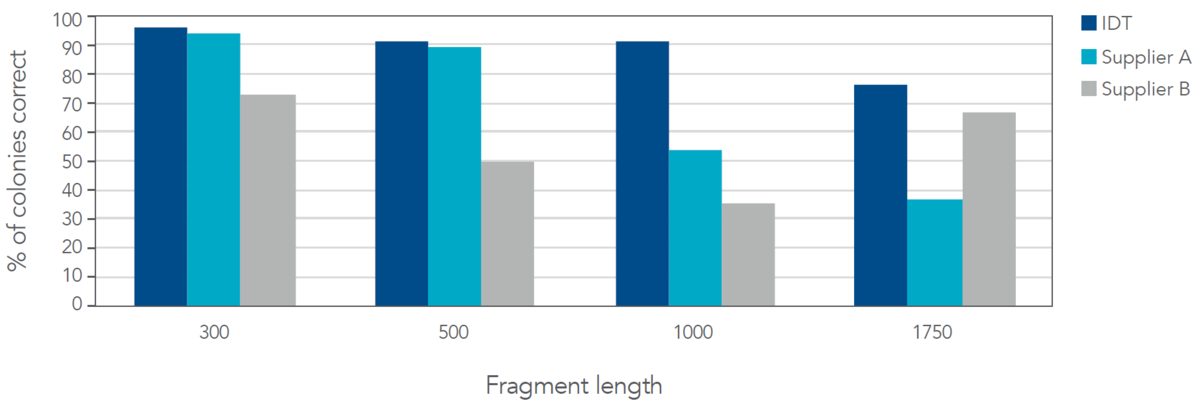
Figure 5. IDT gene fragments produce a higher percentage of correct colonies when compared to two other suppliers. Based on screening and sequencing of 24 colonies for each sequence, IDT dsDNA fragments were the only fragments to have greater than 75% of correct colonies selected that contained the desired full-length sequence and were also the only fragments to provide greater than 90% correct colonies selected for fragments 1 kb or less in length.
At all lengths, IDT fragments outperformed those from Supplier B. In some cases, nearly a two-fold increase in correct sequences was obtained. IDT and Supplier A performed similarly at smaller length ranges; however, IDT fragments significantly outperformed those from Supplier A at lengths 1 kb and above.
Conclusion
This head-to-head comparison against two other major suppliers demonstrates how the IDT proprietary synthesis, assembly, and correction processes result in fragments with lower error rates, in fact, as low as 1:6300. These error rates brought about improved functionality in cloning applications with an improvement in correct clones as high as 100%. This increase could lead to researchers being able to reduce the number of colonies they need to screen by as much as 50%.
By minimizing the number of colonies needed to be screened, IDT gene fragments can reduce the rework steps researchers need to perform due to expression failure. Gene construction should be a seamless step in a research workflow and should not be aggravated by supplier issues such as unwanted mutations and truncations. Researchers using IDT gene fragments for cloning applications can therefore advance their studies rapidly in a budget-friendly manner to meet increasingly shorter project demands.
More than decreased error rates, IDT gene fragments have a short turnaround time and can be shipped in as few as 2 business days from order acceptance. So, as a reliable provider of high-quality dsDNA, IDT supplies gene fragments ideal for setting up cloning experiments for a variety of length ranges.
Download
Who is IDT?
Integrated DNA Technologies (IDT) is your advocate for the genomics age. They produce tools for NGS, CRISPR, qPCR and PCR. Their products include DNA/RNA oligos, genes and gene fragments. For more than 30 years, IDT's innovative tools and solutions for genomics applications have been driving advances that inspire scientists to dream big and achieve their next breakthroughs.