Glycosylation and Methylation
The complexity of the human proteome far outweighs that of the human genome, largely due to post-translational modifications (PTMs). Previously, we looked at three of the most widely studied PTMs: phosphorylation, ubiquitination, acetylation. Here, we focus on two other major PTM types: glycosylation and methylation.
Glycosylation
Glycosylation is the covalent attachment of single sugars (monosaccharides) or glycans (oligosaccharides and polysaccharides) to select residues of target proteins. Because multiple aspects of glycosylation can vary, including the type of sugar, as well as glycan length, composition, and structure, glycosylation is thought to increase the diversity of the proteome more than any other PTM. Glycosylation can influence protein folding, conformation, stability, solubility, distribution, and activity. It has roles in biological processes including cell adhesion, receptor activation, molecular trafficking, signal transduction, and cell–cell / cell–matrix interactions. Glycosylation is classified into six main groups based on the residues that are targeted:
N-glycosylation
N-glycosylation occurs co-translationally at the carboxamido nitrogen of asparagine (N) residues. It is initiated in the lumen of the endoplasmic reticulum (ER) and driven by a membrane associated enzyme complex called the oligosaccharyltransferase (OST), which is involved in transferring preassembled blocks of 14 sugars (including 2 N-acetylglucosamines, 9 mannoses and 3 glucoses) to the nascent polypeptide chain1. The protein is then transferred to the Golgi apparatus for further processing (terminal glycosylation).
O-glycosylation
O-glycosylation occurs post-translationally on amino acids with functional hydroxyl (-OH) groups, which are predominantly serine and threonine side chains or, to a much lesser extent, hydroxyproline and hydroxylysine residues. Unlike N-glycosylation, the bulk of O-glycosylation takes place in the Golgi apparatus after the protein has been folded, with the monosaccharides being added sequentially by a multitude of different glycosyltransferase enzymes2.
C-glycosylation
C-linked glycosylation involves the formation of a carbon-carbon bond to link a mannose residue to a tryptophan (W) residue within an extracellular protein. It occurs in the ER and is driven by the action of C-mannosyltransferase enzymes, which attach monomeric α-mannose to the first tryptophan in the (W-x-x-W/C) motif of target proteins3.
S-glycosylation
S-linked glycosylation refers to the conjugation of a sugar to the sulfur atom (S) of a cysteine residue. While extremely rare in comparison to other types of glycosylation, S-glycosylation is of interest to researchers as it is known to yield glycopeptides such as sublancin, an antimicrobial natural product4,5
P-glycosylation
P-glycosylation, also known as phosphoglycosylation, is the attachment of a phospho-sugar to a serine or threonine residue. To date, it has only been observed in slime molds and a small number of unicellular parasites, but many other examples of P-glycosylation are thought to exist6.
Glypiation
Glypiation describes the attachment of a glycosylphosphatidylinositol (GPI) anchor to the C-terminus of a newly-synthesized protein. It is carried out by an ER-situated transamidase and serves to localize proteins to cell membranes. GPI-anchored proteins have diverse functions, including roles in embryogenesis, neurogenesis, the immune response, and signal transduction7. Defects in the genes involved in their synthesis and processing have been linked to a broad array of disorders.
Methylation
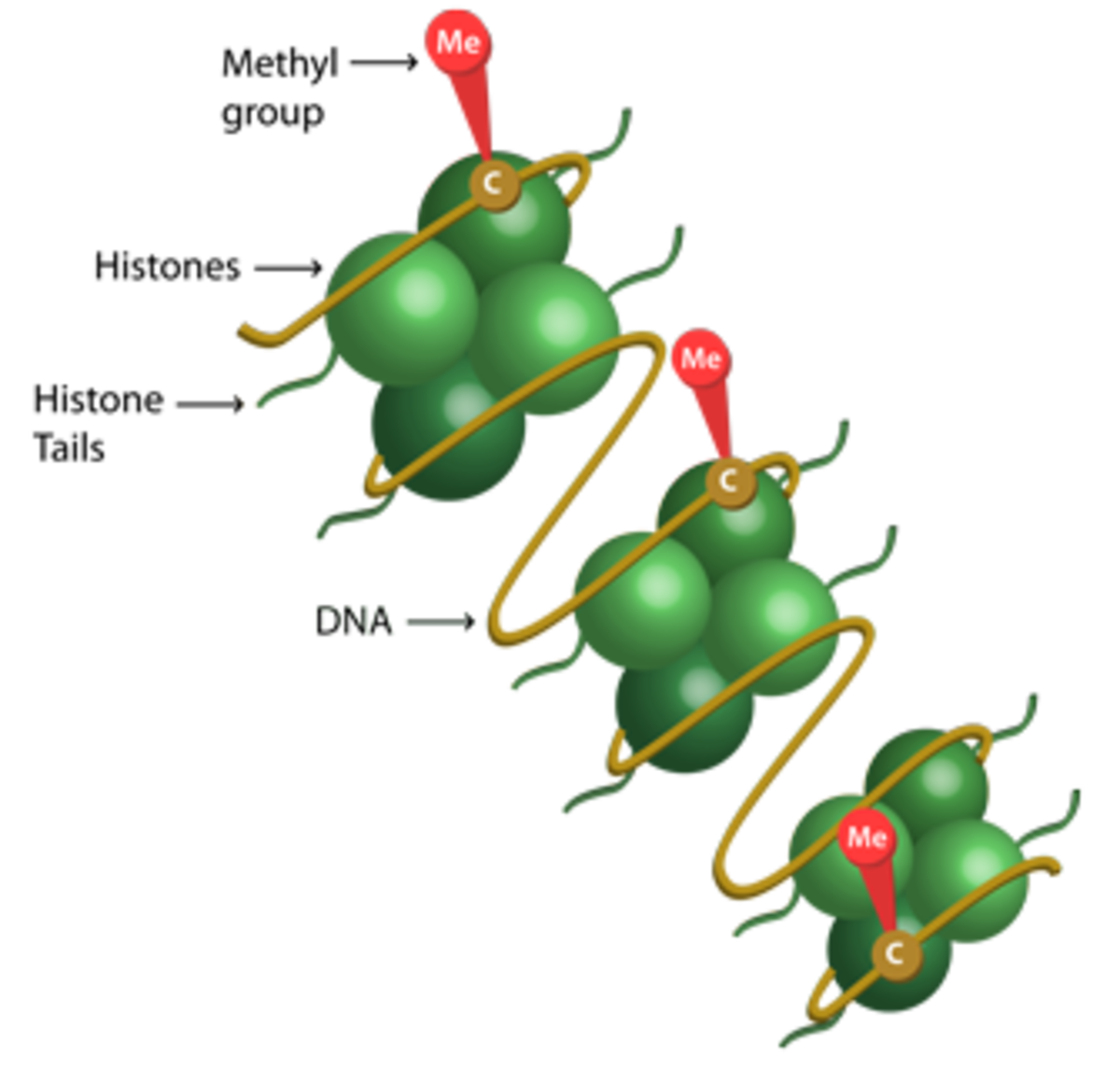
Methylation involves the transfer of a methyl (CH3) group to a target protein. It is best known for its role as a histone PTM, which occurs mainly at specific lysine (K) and arginine (R) residues under the control of histone methyltransferase (HMT) and histone demethylase (HDM) enzymes to regulate gene expression.
In mammals, the canonical lysine methylation sites are found on histone H3 at lysine 4 (H3K4), lysine 9 (H3K9), lysine 27 (H3K27), lysine 36 (H3K36) and lysine 79 (H3K79), and on histone H4 at lysine 20 (H4K20)8. The most extensively studied arginine methylation sites in histones include H3R2, H3R8, H3R17, H3R26, and H4R39.
While lysine residues offer three possible forms of methylation, mono-methyl, di-methyl, and tri-methyl, arginine methylation can be either mono-methyl, symmetric di-methyl, or asymmetric di-methyl10. In combination, the methylation site and type of methylation exert different downstream effects.
Other types of proteins that undergo methylation include the Ras proteins, the lamins, and various kinase enzymes, all of which perform different essential functions.
LubioScience represents some of the most trusted brands in research and works closely with our partners to offer high-quality reagents for studying PTMs including glycosylation and methylation. Contact us today to discuss how we can support your project.
Refrences
- https://pubmed.ncbi.nlm.nih.gov/32316603/
- https://pubmed.ncbi.nlm.nih.gov/36214107/
- https://pubmed.ncbi.nlm.nih.gov/34500691/
- https://pubmed.ncbi.nlm.nih.gov/21196935/
- https://pubmed.ncbi.nlm.nih.gov/30582697/
- https://pubmed.ncbi.nlm.nih.gov/9451009/
- https://pubmed.ncbi.nlm.nih.gov/30054924/
- https://pubmed.ncbi.nlm.nih.gov/33859667/
- https://pubmed.ncbi.nlm.nih.gov/31252653/
- https://pubmed.ncbi.nlm.nih.gov/36639369/
