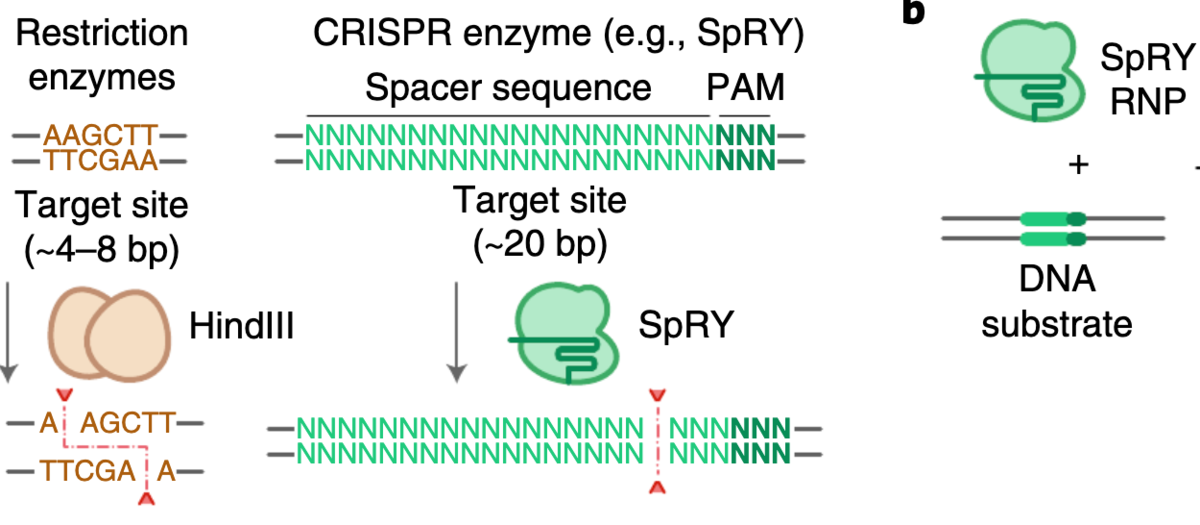
Researchers from Massachusetts General Hospital have engineered a new CRISPR-Cas variant which no longer need a protospacer adjacent motif (PAM) and is able cut at any DNA base in vitro. Its ability to make precise breaks anywhere could be applied to a number of DNA engineering applications.
Typically, wild-type CRISPR Cas9 or Cas12 nucleases require a PAM near the target site, which limits where the enzymes may cleave. The novel SpRY Cas variant, by contrast, has significantly more relaxed requirements. Writing in Nature Biotechnology, Kleinstiver and colleagues report that SpRY can make cuts at nearly any DNA sequence in vitro as long as a guide RNA is present.
The researchers first compared the ability of wild-type SpCas9 and SpRY to create double-stranded breaks at 20 different target locations along a DNA substrate with varying PAM motifs. Using 20 different guide RNAs, they were able to show that wild-type Cas9 could digest four of the sites to near completion, whereas SpRY managed do so for 19 of the sites.
They also compared the activity of SpRY with another Cas9 variant called SpG, which was engineered to target only certain PAMs. Their findings underscore SpG's preference for NGN, limiting its ability to fully cut the 64 sites tested. SpRY, on the other hand, was able to digest 59 of the 64 sites to near completion, suggesting it is indeed nearly PAMless.
The novel SpRY may be used in a wide range of applications, such as isothermal assembly in molecular cloning, construction of saturation mutagenesis libraries, to deplete unwanted sequences from next-generation sequencing libraries, or in target enrichment in sequencing protocols.
SpRY is straightforward to use, as the authors note. The oligonucleotides can be purchased from any vendor, the gRNA transcription kits are already commercially available, and the SpRY protein will soon be available from vendors as well. Kleinstvier hopes that the simplicity of his method will permit users to incorporate SpRY into typical molecular cloning workflows without too many challenges.