What is Next Generation Sequencing?
First launched at the beginning of the 21st century, Next Generation Sequencing (NGS) has provided massive parallel sequencing of DNA and RNA genomes. Like Sanger sequencing, NGS starts with fragmented DNA or RNA, which is followed by the addition of sequencing adapters. Then labelled nucleotides are added one at a time to a growing DNA template strand to sequence and analyze the data. However, unlike Sanger technology, which sequences one DNA fragment at a time using capillary electrophoresis, NGS allows massive parallel sequencing using solid surface technology.
NGS methods are highly efficient, and different genomic features, including single nucleotide variants (SNVs), copy number, structural variants and RNA fusions, can be analyzed. The parallel sequencing offered by NGS makes the technology faster and more cost-effective over Sanger sequencing or qPCR methods when analyzing more than 20 target regions or high sample volumes. This makes NGS a popular choice for research and clinical laboratories.
LubioScience work with the global leader in oligonucleotide synthesis, Integrated DNA Technologies (IDT), to provide a range of ready- or custom-made solutions for your NGS applications. Sequencing by synthesis, employed by Illumina, is the most widely adopted sequencing technology. So, here we focus on the different NGS adapters and indexing primers available for Illumina sequencing.
What are NGS adapters?
Adapters are key components for NGS as they act in the first step in a NGS workflow: library preparation. Adapters are short pieces of DNA around 80 bases in length, which attach to the DNA fragments of interest to be combined with primers for amplification. Adapters also bind to the DNA linkers on the flow cell's solid surface, allowing the NGS workflow's sequencing stage to occur.
General structure of the NGS adapter
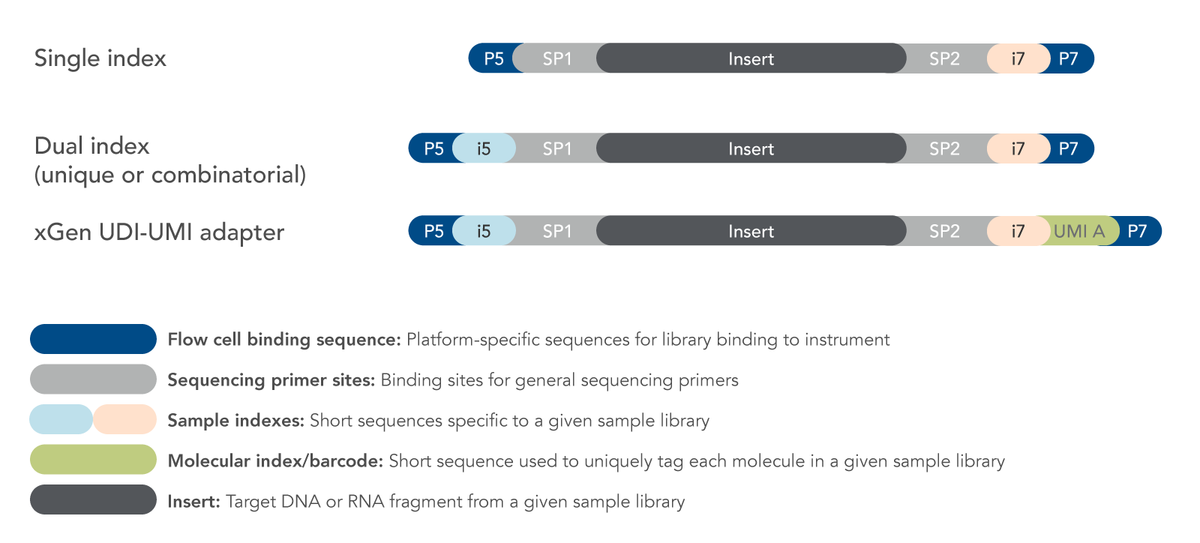
Adapters flank either side of the sequence of interest. Essentially, there are three main components in each NGS adapter. The first is the flow cell binding sequence, which is platform-specific and used to bind the flow cell. The flow cell is where the sequencing occurs. For binding to the flow cell, adapter oligos are complementary to the oligos attached to the sequencing flow cell. The P5 adapter binds to the 5'end of flow cell oligos, and the P7 adapters bind to the 3'end.
Another part of the NGS adapter molecule is the sequencer primer binding site. This region allows the binding of the sequencing primers, which consequently enables the recruitment of the polymerases to bind and extend the oligo synthesis. Single-end sequencing reads a sequence from one end to the other. In comparison, pair-end sequencing allows the sequencing of both ends of a fragment.
NGS Adapters can also have tags — like sample barcodes. This tagged region of the adapter is called an index or barcode region. This part of the molecule allows hundreds of samples to be pooled and sequenced in a single run without getting muddled up — sample multiplexing.
Different types of indexing exist. Deciding on the indexing strategy depends on several factors. These include the input sample, the number of samples and target regions, the level of multiplexing needed, the desired speed of processing, and the need to mitigate crosstalk or sample bleeding.
Single index sequencing
Single-index (SI) sequencing generally consists of 8–10 bases and is specific to a single library. Their purpose is to simplify downstream analysis by assigning individual sequence reads to the correct sample.
SI adapters contain a 6-base index 1 (i7) sequence used in the downstream analysis to identify and separate the sample libraries. This i7 index read follows read 1, which is also called the "forward read". Read 1 extends in the 5’-3’ direction along the forward DNA strand. Single index adapters allow the addition of up to 48 unique 6-base i7 sequences, thus generating up to 48 uniquely-tagged libraries.
SI adapters are ideal for workflows requiring fast turnaround times and processing rates, as only the single i7 Index needs to be read. Example workflows are those that do not need library multiplexing or require high resolution. Run times employing SI are shorter than dual index sequencing, which uses both Index 1 and 2 reads.
Dual-index sequencing
Dual-index (DI) sequencing contains two index sequences, i7 and i5. The index read 1 (i7 adapter) is read directly after read 1. Index read 2 (i5 adapter) is read either before or after read 2 resynthesis, depending on the workflow and system used for the dual-index workflow on a paired-end flow cell. Read 2, also known as the "reverse read" extends in the 5’-3’ direction along the reverse DNA strand.
DI adds an additional step to avoid muddling up samples to increase the accuracy of read identification of the sequencing data. The two types of DIs are detailed below.
- Unique dual indexing (UDI)
One type of DI is unique dual indexing (UDI). UDI is primarily used to avoid a phenomenon known as index hopping – incorrectly assigning the expected Index to a different Index from a multiplexed pool – which can reduce data quality. Using UDIs allowed hopped reads to be removed from the downstream analysis as unexpected combinations will be removed from the data. One UDI plate consists of 96 unique Index 1 (i7) adapters and 96 unique index 2 (i5) adapters. This avoids the unambiguous assignment of indexes to libraries as samples are labelled. UDI adapters are ideal for applications that require high sensitivity. Examples include detecting rare transcripts and identifying the presence or absence of low-abundance strains.
- Combinatorial dual indexes
The other type of DI is called combinatorial DI. As the name suggests, combinations of indexes are employed. Each i7 and/or i5 Index is repeated across the rows and columns of a well plate. Using the different i7 and i5 adapters multiple times in different combinations
significantly increases the multiplexing capacity of a sequencing lane and run. CDI cannot easily identify hopped Index reads. However, a certain level of sample verification exists. Only data from i7 and i5 matching the expected combinations can be considered in the downstream data analysis. CDI is suited to applications tolerant of low levels of misassignment between samples.
Unique molecular identifiers (UMIs)
NGS Adapters can also contain sample-specific identifiers known as unique molecular identifiers (UMIs). UMIs differ from library-specific DIs. UMIs are short random nucleotide sequences positioned next to the i7 Index, so an UMI is sequenced as part of the i7 Index read.
UMIs adapters help increase variant identification by reducing the rate of false-positive variant calls. UMIs set apart true variants from errors caused during library preparation, target enrichment or sequencing. UMIs also identify PCR duplicates, as the duplicates would contain the same insert and UMI tag. Other UMI applications are RNA-seq gene expression analysis and other quantitative sequencing methods.
Adapters are also available with UDI and UMI, such as the IDT xGen UDI-UMI indexes or custom-made adapters with these configurations. Located next to the i7 Index, the UMI is sequenced as part of the i7 Index read. Research has shown that using UDI-UMI adapters can improve the accuracy of quantitative NGS.
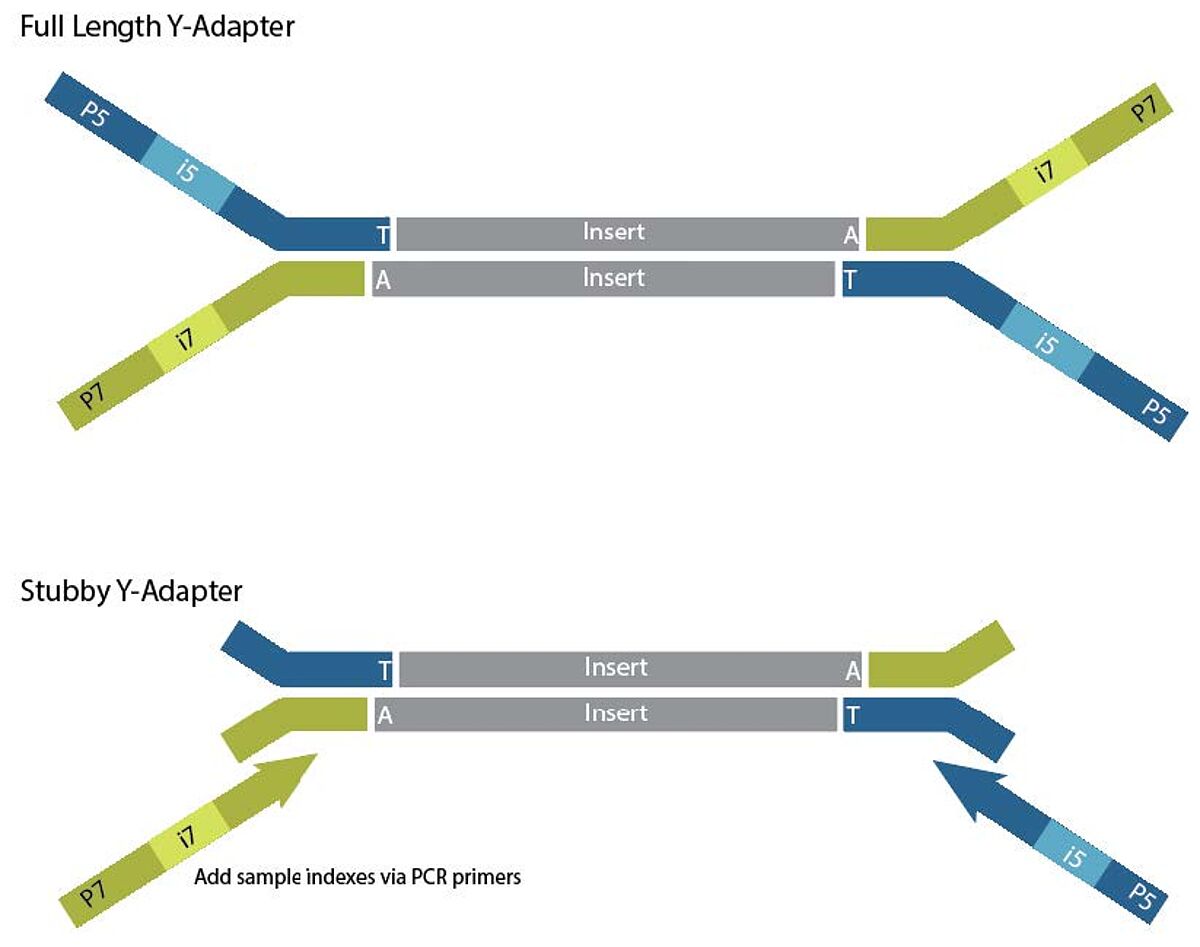
Adapter designs and indexing primers
Illumina workflows employ two types of adapter designs: full-length and Stubby-Y adapters. Full-length adapters are ligated to A-tailed library fragments. Stubby adapters are ligated to libraries generated with TA ligation. If using protocols with Stubby adapters, indexing primers with UDI primer pairs are also needed for indexing PCR. Indexing primers contain unique i5, i7 indexes and P5 and P7 sequences. Stubby adapters and indexing primers are used for various applications, from whole-genome sequencing to targeted sequencing for library preparation, where indexing with PCR is preferred. Indexing primers are also available for Nextera workflows based on tagmentation and PCR. Truncated adapters and indexing primers offer higher DNA ligation efficiencies than full-length adapters. However, a PCR step is needed to add the indexing primers to the ligated Stubby adapters, meaning Stubby adapters and indexing primers cannot be used with PCR-free libraries.
LubioScience's NGS adapters
Using high-quality NGS adapters is key to producing high-quality sequencing data. IDT's NGS adapters and indexing primers, available from LubioScience, use state-of-the-art proprietary processes (TruGrade™) to synthesize and purify NGS adapters. Sample indexes are compatible with 2- and 4-color sequencing platforms. LubioScience's catalog also includes methylated adapters for bisulphite sequencing (Methyl-Seq). LubioScience also work with IDT to design and produce custom adapters for your specific application.
Please do not hesitate to contact us for further assistance in choosing the suitable solution for your application needs.
Please visit our website for more details. Contact us to learn more about NGS adapters.
IDT
Integrated DNA Technologies (IDT) is your advocate for the genomics age. They produce tools for NGS, CRISPR, qPCR and PCR. Their products include DNA/RNA oligos, genes and gene fragments. For more than 30 years, IDT's innovative tools and solutions for genomics applications have been driving advances that inspire scientists to dream big and achieve their next breakthroughs.