Selecting competent cell for your application needs
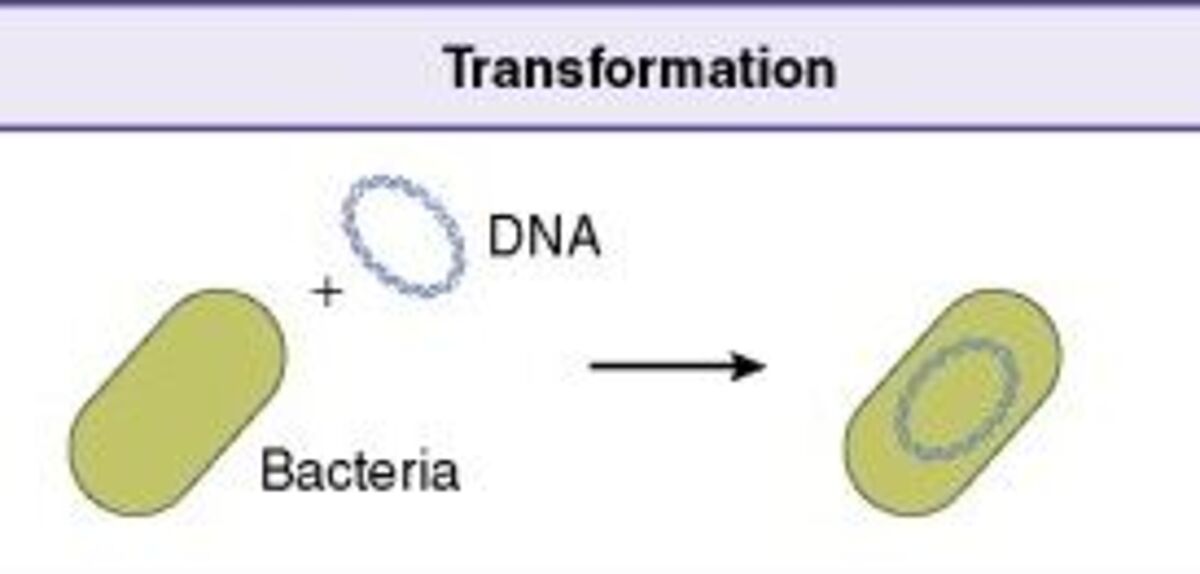
Bacterial transformation is a horizontal gene transfer process that allows certain bacteria to naturally uptake genetic material from the environment, known as natural competence. Bacterial transformation has allowed bacteria to evolve by responding to selective pressures and adapting to new environments. Bacteria can be made competent and permeable to DNA, known as artificial competence. Molecular cloning techniques have taken advantage of artificial transformation to insert recombinant DNA into host cells.
Different preparations of competent cells
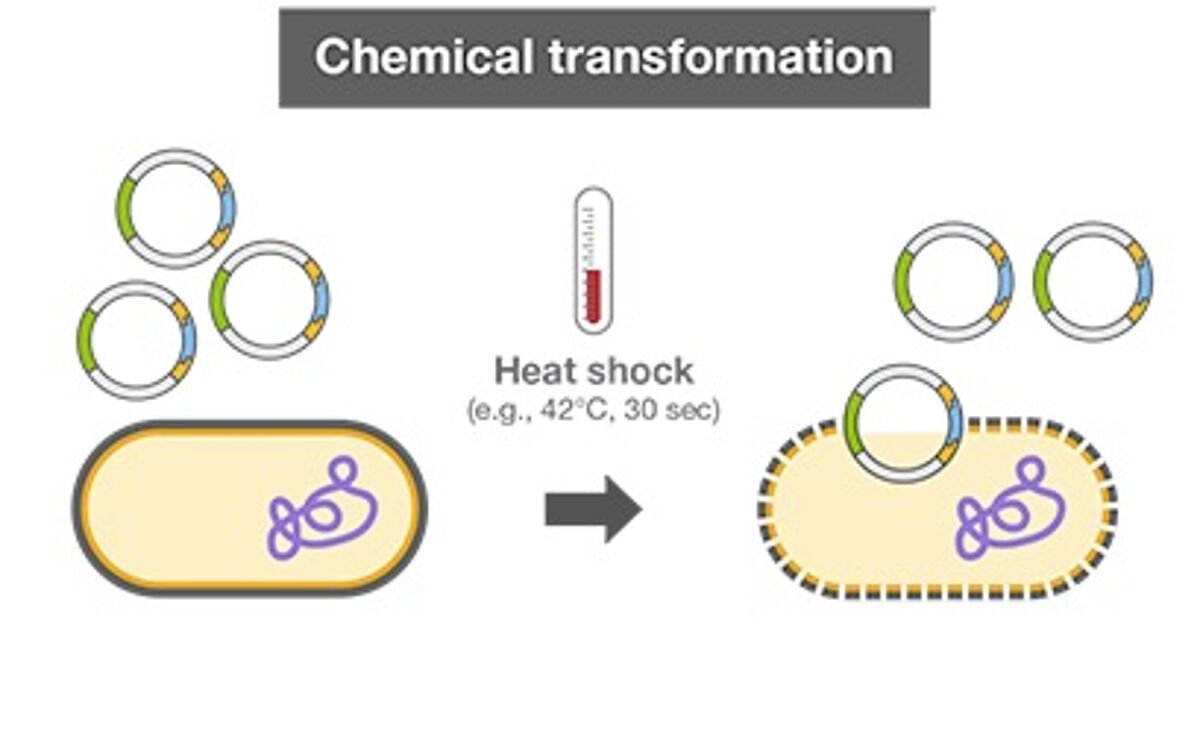
Chemical transformation and electroporation are the two main protocols used to introduce DNA into competent bacterial cells. Both methods involve growing and harvesting bacterial cells. Chemical transformation involves treating bacterial cells with calcium chloride solution to disrupt the cell membrane. After a brief heat shock treatment, DNA can enter the chemically treated bacterial cells. The success of the transformation reaction (or transformational efficiency) is defined by how successfully competent cells receive additional genetic material and express genes encoded by it. Cell viability can vary greatly in chemically treated cells, which affects transformation. Premade competent cells have minimal batch-to-batch variability and also save time on protocols.
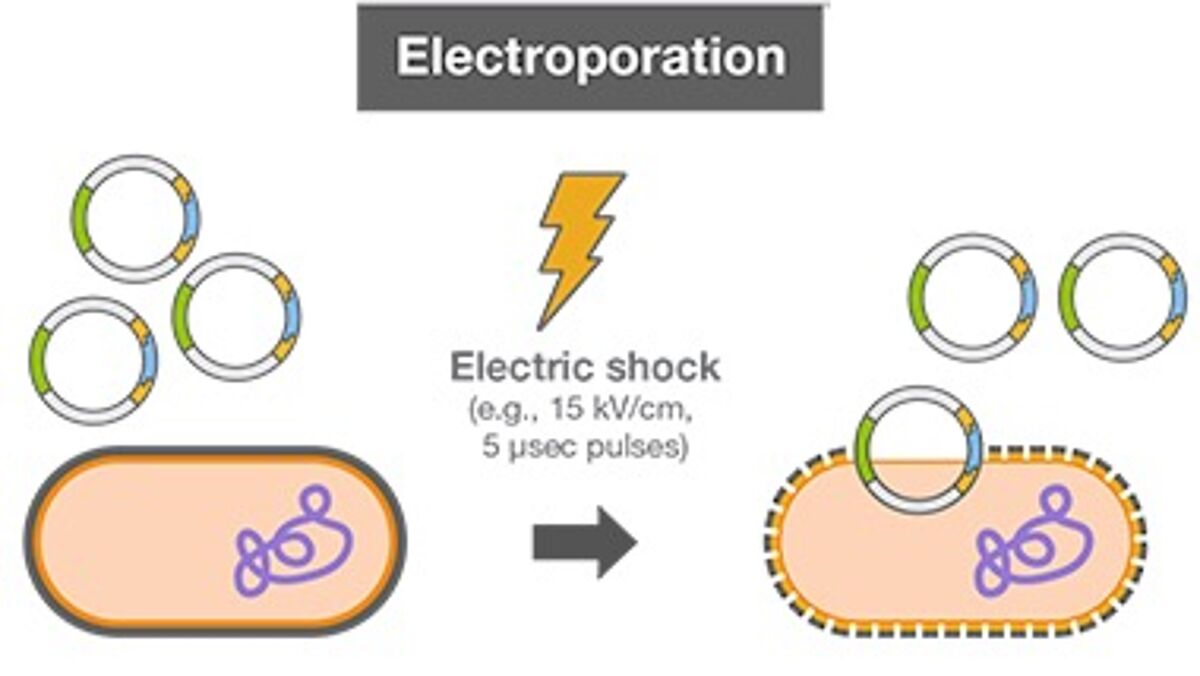
Electroporation - Another way to introduce DNA into competent cells is by exposing cells to an electric field. After harvesting bacterial cells, DNA and cells are inserted into cuvettes in an electroporation instrument and subjected to an electrical current, which allows the DNA to enter the cells. Electrocompetent cell preparation involves numerous washing steps to remove salts from the media the cells were grown in, as salts hamper the electroporation reaction. Commercial electrocompetent cells have high transformation efficiencies.
LubioScience distribute a wide range of ready-to-use chemically competent and electrocompetent cells from Biosearch Technologies. For both methods of transformation, a nutrient-rich growth medium is needed after transformation to help bacterial cells recover. Control DNA assesses the transformation reaction. The recovery medium and control DNA are supplied with LubioScience's competent cells. To choose the best competent cell option, it is essential to consider the bacterial strain of the competent cell, the cloning vector and the transformation method.
Choosing competent cell strains depending on the application
Different gene technology applications include routine cloning, generating complementary DNA (cDNA) libraries and CRISPR single guide RNA (sgRNA) libraries. The type of competent bacterial cells needed is dependent on the application. Generally, routine cloning procedures use chemically competent cells, and electrocompetent cells are ideal for cloning larger fragments and library construction. Below, we describe how certain genetic markers in the genotype of competent bacterial cells determine their applicability in experimental protocols.
Selection and screening methods used for most transformation protocols, including routine cloning
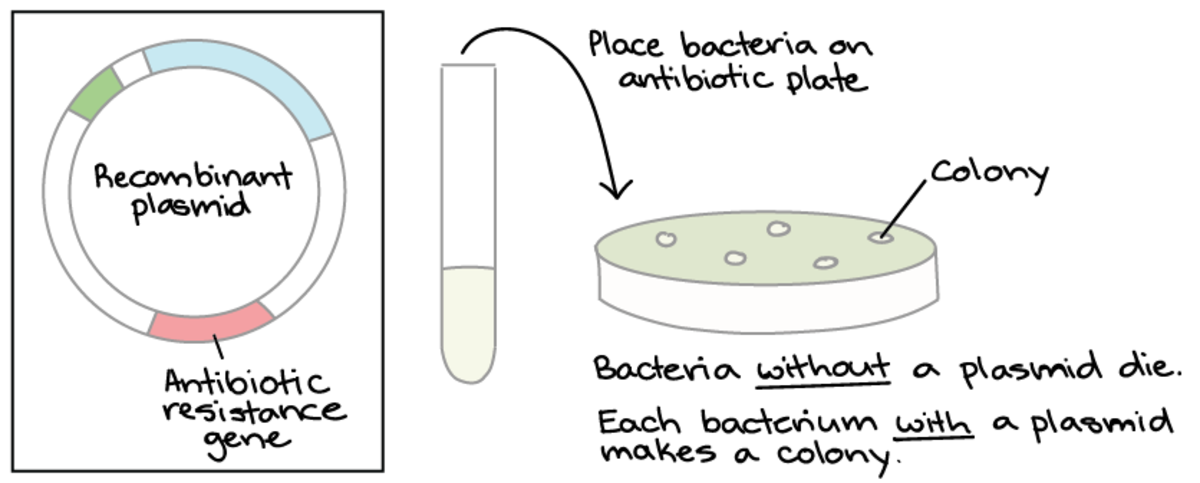
Selectable markers are genes introduced into a cell that confer an ability to select positively transformed cells from the wildtype. Competent bacterial cells lacking antibiotic resistance are employed to screen for the inclusion of recombinant DNA carrying antibiotic-resistance genes when grown on growth media containing antibiotics (selective media). Antibiotic selective markers employ antibiotics commonly found in laboratories, including ampicillin, streptomycin, tetracycline, gentamicin and chloramphenicol. Competent cells only grow and form colonies on selective media when the insert has successfully ligated into the vector, and the DNA has been taken up by competent cells.
Resistance to lytic bacteriophages like bacteriophage T1 is important as bacterial host cells are lysed and die after infection with lytic phages. Bacterial strains with a mutation in tonA (now called fhuA) gene prevent bacteriophage T1 from binding and infecting cells (Rostøl & Marraffini, 2019). Several of LubioScience's competent cells are resistant to T1 phage infection.
Blue-white screening is s commonly used in cloning applications. Bacterial cells containing mutations in the lac operon (lacZΔM15) cannot metabolise lactose or lactose analogues. If transformation occurs with recombinant DNA containing a short segment of the lacZ gene, a functional β-galactosidase enzyme can be reconstituted. When a lactose analogue, X-gal, is present in the solid growth media, functional β-galactosidase can hydrolyse X-gal and produce a blue dye, which results in blue colonies. When a DNA insert has successfully ligated into the vector to produce recombinant clones, lacZ coding from the vector is disrupted, resulting in white colonies. This makes blue-white screening an important tool in bacterial colony selection.
LubioScience has several competent cells for routine cloning, such as E. cloni 10G, 10F' and 5-alpha cell range. Available in chemically competent and electrocompetent formats, these cells are direct replacements for common cloning strains (e.g., DH10B, Top10, DH5alpha, XL1-Blue).
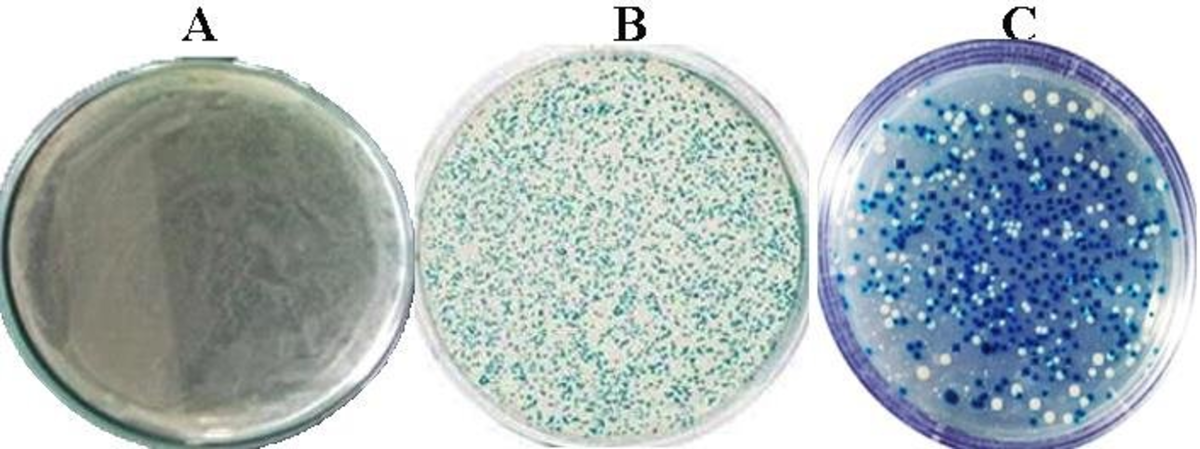
Transformation and screening results of the blue and white colonies from E.coli recombinant.
A. (negative control), E. coli JM109 competent cells without plasmid. B (positive control) E coli cells growth where blue colony containing plasmid pGEM-T Easy without insertion DNA transformed, and C. E coli cells growth where white colony contains pGEM-T Easy-MPT64 recombinant plasmid and theblue colony is self-ligated.
(Ahmad, 2019)
Cloning large inserts
Cloning large inserts is challengingas transformation efficiency decreases as plasmid size increases (Leonardo & Sedivy, 1990; Siguret et al., 1994). Mutations in the recA recombination gene reduce recombination with plasmids and chromosomal DNA and result in greater stability of large cloned inserts. Mutations in the endA1 gene also eliminate the non-specific digestion of plasmid DNA. Therefore, endonuclease minus and recombination minus competent bacterial strains improve the quantity and quality of plasmid DNA and increase the cloning efficiency of large plasmids and ligated DNA. The TransforMax range of competent cells is optimised for cloning large inserts, plasmids and fosmids.
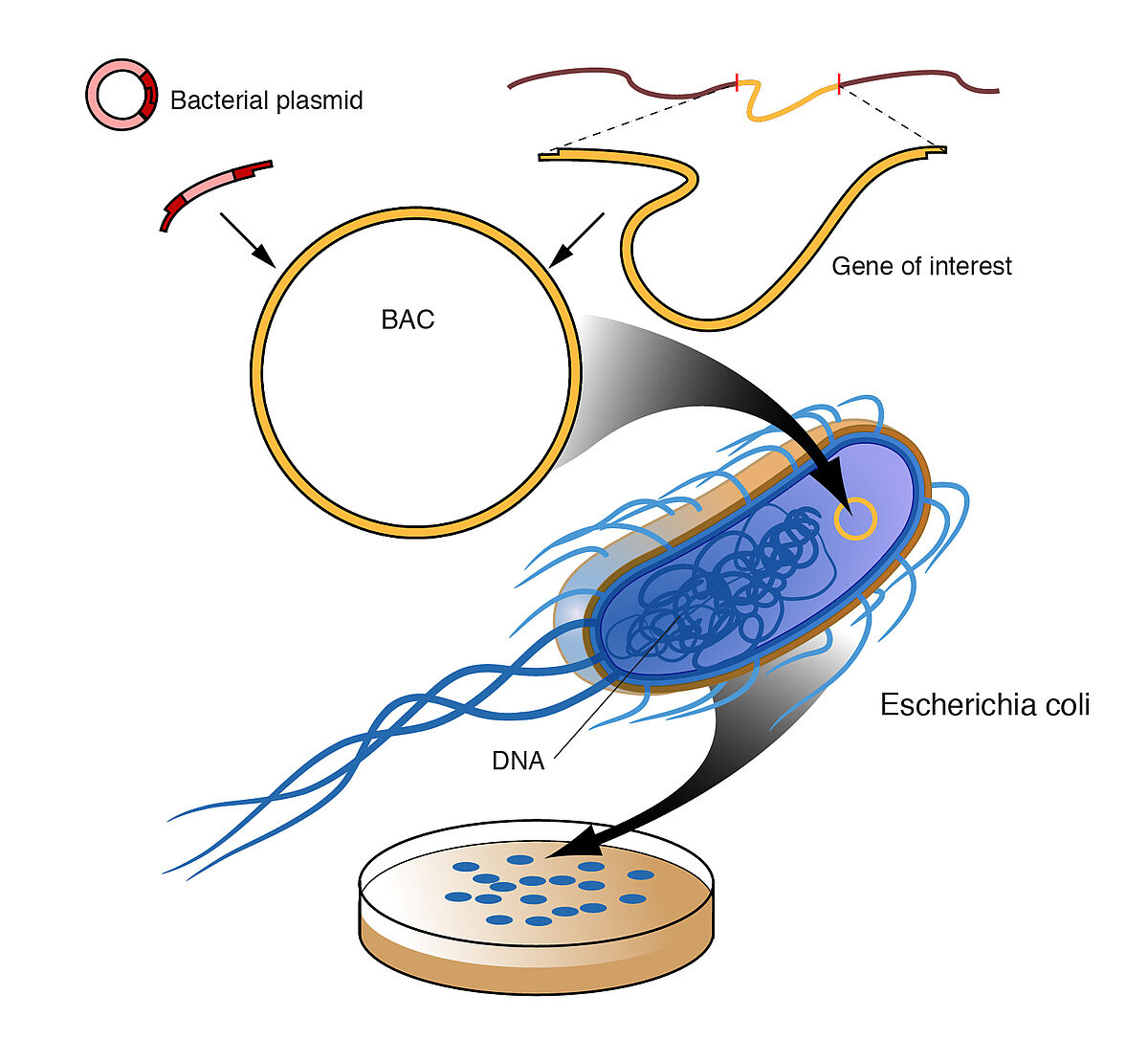
The endA1 mutation is also present in competent bacterial cells that are optimised for cloning and propagating the large size of DNA inserts in Bacterial Artificial Chromosomes (BAC). The BAC-Optimized Replicator™ v2.0 Electroporation Competent Cells also contains an arabinose-inducible trfA gene, which is required for induction of replication from the oriV origin in BAC and fosmid clones. LubioScience provide BAC-Optimised Electrocompetent Cells that result in high yields of high-quality BAC/plasmid DNA.
Cloning methylated DNA
Cloning highly methylated eukaryotic DNA can be hampered by bacterial hosts cleaving the methylated sequences using restriction modification systems as they are methylated in a different way to that of the host. Using competent cells deficient in endogenous restriction systems increases the efficiency of introducing methylated DNA into bacterial cells. LubioScience's E. cloni 10G and 10F' cell range are restriction-deficient strains ideal for routine cloning of methylated DNA. The TransforMax strains are restriction-deficient strains that improve the size and representation of libraries constructed with methylated DNA.
Other applications
LubioScience also offer competent cells ideal for constructing phage display libraries and protein expression (TG1, ER2738, SS320, MC1061F' and MC1061), as well as preparation of single-stranded DNA (ssDNA) site-directed mutagenesis with Kunkel's method (CJ236).
Genotypes of LubioScience's competent cell range
Name | Genotype |
General cloning and library construction | |
(Electrocompetent and chemically competent) | F¯ mcrA ∆(mrr-hsdRMS-mcrBC) endA1 recA1 φ80dlacZ∆M15 ∆lacX74 araD139 ∆(ara,leu)7697 galU galK rpsL (StrR) nupG λ− tonA |
(Electrocompetent) | [F' proA+B+ lacI q ZDM15::Tn10 (TetR)] / mcrA D(mrr-hsdRMS-mcrBC) endA1 recA1 f80dlacZDM15 DlacX74 araD139 D(ara,leu)7697 galU galK rpsL (StrR ) nupG l- tonA |
(Chemically competent) | fhuA2Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 |
Unstable Inserts or Toxic Gene Products | |
(Electrocompetent and chemically competent) | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ- rpsL (StrR) nupG trfA tonA pcnB4 dhfr |
Large Inserts, Plasmids, or Fosmids | |
(Electrocompetent and chemically competent) | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ- rpsL (StrR) nupG |
(Electrocompetent and chemically competent) | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ- rpsL (StrR) nupG trfA dhfr |
(Electrocompetent) | F - mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ - rpsL (Str R) nupG trfA tonA |
BAC Cloning | |
BAC-Optimized Replicator v2.0 Genotype (Electrocompetent) | F¯ mcrA Δ(mrr-hsdRMS-mcrBC) endA1 recA1 Φ80dlacZ ΔM15 ΔlacX74 araD139 Δ(ara,leu)7697 galU galK rpsL (StrR) nupG (attL araC-PBADtrfA250 bla attR) λ¯ |
(Electrocompetent) | F - mcrA Δ(mrr-hsdRMS-mcrBC) endA1 recA1 Φ80dlacZΔM15 ΔlacX74 araD139 Δ(ara, leu)7697 galU galK rpsL nupG λ- tonA (StrR) |
(Electrocompetent) | F- mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 endA1 recA1araD139 Δ(ara,leu)7697 galU galK rpsL nupG λ- tonA AmpR sopAB telN antA |
Phage Display | |
(Electrocompetent) | [F' traD36 proAB lacIqZ ΔM15] supE thi-1 Δ(lac-proAB) Δ(mcrB-hsdSM)5(rK - mK -) |
(Electrocompetent) | F'[proAB lacIqZ ΔM15 Tn10 (TetR)] araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (StrR) hsdR2 (rK– mK+) mcrA mcrB1 |
(Electrocompetent) | [F'proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10 (tetr)] fhuA2 glnVΔ(lac-proAB) thi-1Δ(hsdS-mcrB)5 |
(Electrocompetent) | F-, araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL (StrR) hsdR2 (rK– mK+) mcrA mcrB1 |
CRISPR sgRNA Libraries and Lentiviral Plasmid Cloning | |
(Electrocompetent and chemically competent) | recA13 supE44 ara-14 galK2 lacY1 proA2 rpsL20(StrR) xyl-5 λ– leu mtl-1 F– mcrB mrr hsdS20(rB–, mB–) |
Site-Directed Mutagenesis | |
CJ236 (Electrocompetent) | dut1,ung1,thi-1,recA1 / pCJ105 (F' camr ) |
More information
Contact us
We have an extensive portfolio of competent bacterial cells (Table 1) that are optimised for a wide range of applications, from routine to difficult cloning and protein expression. Also, our competent cells are cheaper than equivalent products from competitors. For more information about choosing a competent cell strain:
References
- Actor (2012) Chap 11 – Basic Bacteriology. Elsevier’s Integrated Review Immunology and Microbiology (Second Edition), 93-103.
- Ahmad, Ahyar & Agus, R & Nasrum, Muhammad & Handayani, I & Karim, Hamidian. (2019). Cloning and characterization of Rv1980c gene encoding MPT64 Protein from Mycobacterium tuberculosis as a new candidate vaccine of tuberculosis. Journal of Physics: Conference Series. 1341. 032010. 10.1088/1742-6596/1341/3/032010. www.researchgate.net/publication/337085421
- Leonardo, E. D., & Sedivy, J. M. (1990). A new vector for cloning large eukaryotic DNA segments in Escherichia coli. Nature Biotechnology, 8(9). doi.org/10.1038/nbt0990-841
- Rostøl, J. T., & Marraffini, L. (2019). (Ph)ighting Phages: How Bacteria Resist Their Parasites. In Cell Host and Microbe (Vol. 25, Issue 2). doi.org/10.1016/j.chom.2019.01.009
- Siguret, V., Ribba, A. S., Cherel, G., Meyer, D., & Pietu, G. (1994). Effect of plasmid size on transformation efficiency by electroporation of Escherichia coli DH5α. BioTechniques, 16(3).
- www.genome.gov/genetics-glossary/Bacterial-Artificial-Chromosome
- www.khanacademy.org/science/ap-biology/gene-expression-and-regulation/biotechnology/a/overview-dna-cloning
- www.thermofisher.com/ch/en/home/life-science/cloning/cloning-learning-center/invitrogen-school-of-molecular-biology/molecular-cloning/transformation/competent-cell-basics