More nutritious plants, precision medicine, healthier animals, and ecosystem transformation – scientists' goals made more attainable using genetic engineering. CRISPR-Cas systems are highly versatile and programmable genome editing tools, sparking scientific breakthroughs in biotechnology and medical research.
Natural CRISPR-Cas systems are prokaryotic immune systems that generate resistance against foreign genetic material, such as bacteriophages and plasmids. CRISPR-Cas utilizes nucleases and noncoding CRISPR RNAs (crRNAs) to recognize and destroy foreign DNA to prevent it from hijacking the host cell.
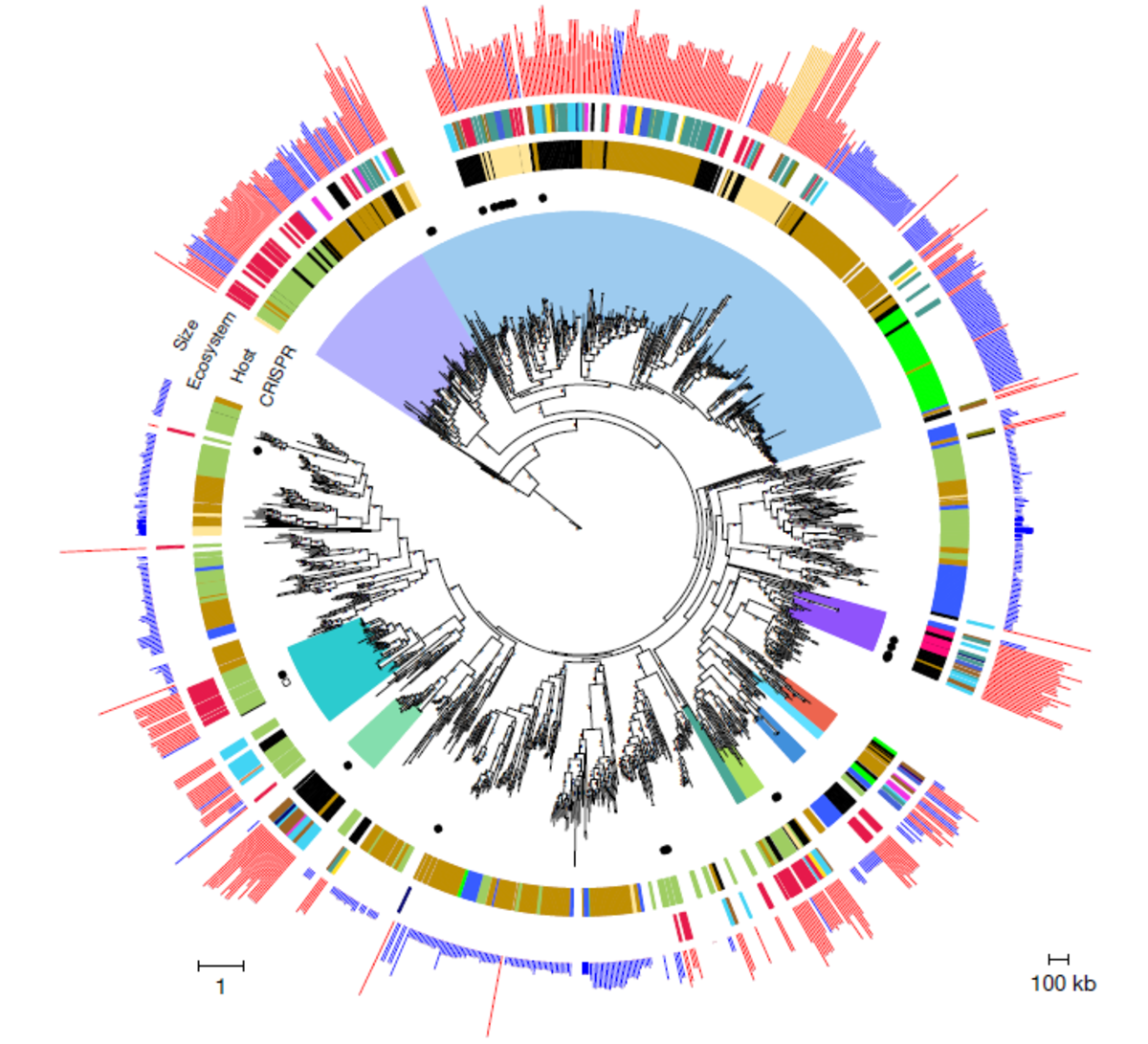
Over the past five years, scientists have leveraged and adapted CRISPR-Cas tools to genetically engineer virtually any living organism by targeting specific regions of the genome. Although they are proven to be useful and powerful tools, they have their limitations, such as off-target effects and lack of predictability, potentially triggering toxic immune responses or unintended outcomes. With the recent discovery of CRISPR-Cas-containing huge bacteriophages (with genomes ranging over 200 kilobases (kb) to 735 kb) being commonly found across a vast array of environments and hosts1, opportunities to expand the genetic engineering toolbox have emerged.
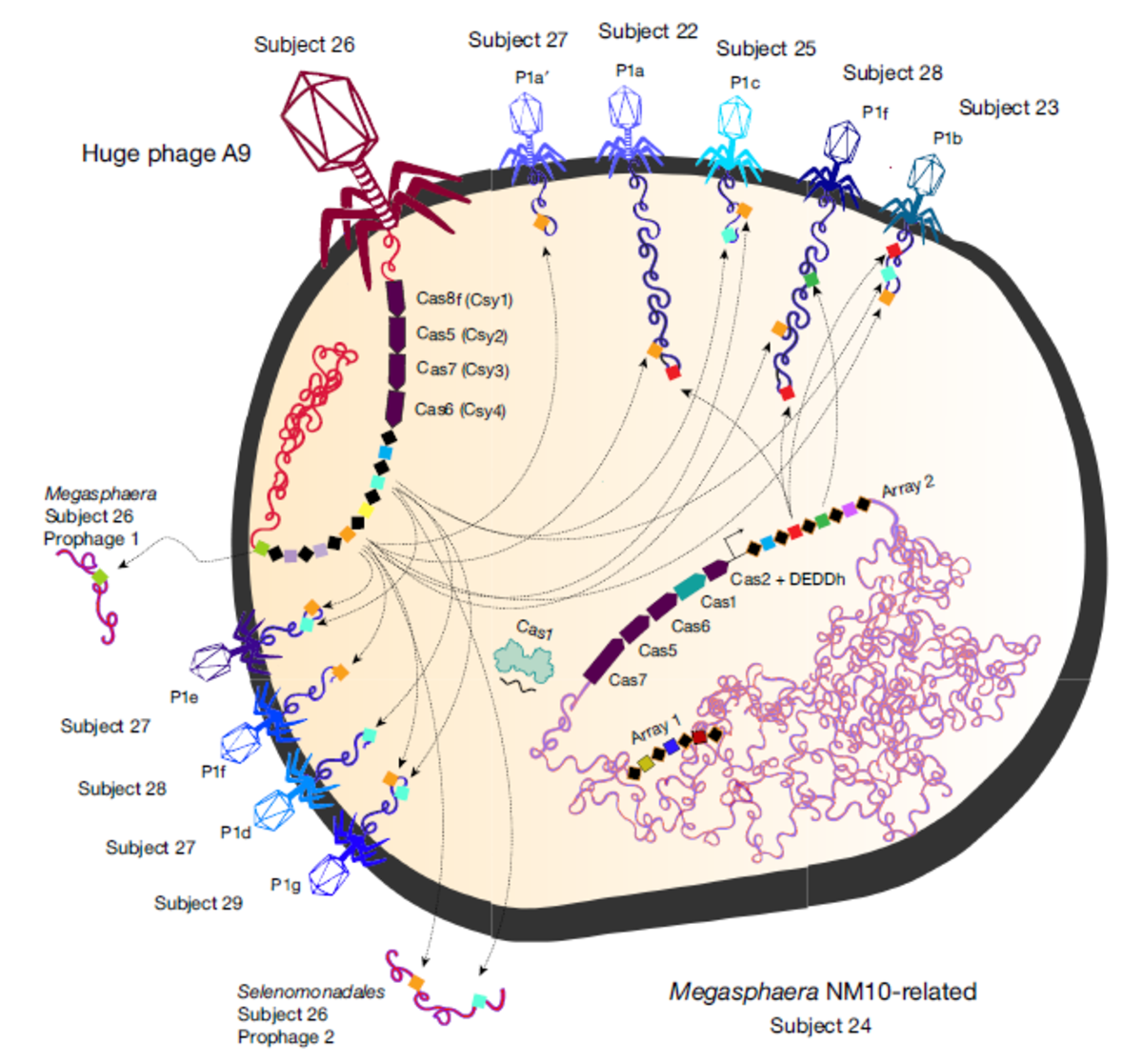
Upon identifying novel CRISPR-Cas systems in huge phages, researchers started exploring what they do and how they work. In a recent publication2, Patrick Pausch, Jennifer Doudna, and colleagues structurally and functionally characterized CasΦ (Cas12j) – a novel Cas protein family encoded in the Biggiephage clade.
Pausch et al. revealed that CasΦ is unusually small with a size of ~70 to 80 kDa – about half the size of Cas9 and Cas12a – and contains a reduced CRISPR array and a nuclease RuvC domain with remote homology to the TnpB nuclease superfamily. With its hypercompact structure, the researchers explored if CasΦ proteins (orthologs CasΦ-1, CasΦ-2, CasΦ-3) could recognize and target DNA. Using E. coli transformation, RNA-seq, and in vitro studies, they demonstrate that CasΦ is capable of cleaving crRNA-complementary DNA, double-stranded and single-stranded, producing staggered 5'-overhangs of 8 to 12 nt – similar to Cas12a. CasΦ-facilitated DNA cleaving required minimal T-rich PAM (protospacer-adjacent motif), a crRNA spacer of ≥14 nt, a mature crRNA, and of course a DNA cleavage active site (RuvC). Surprisingly, CasΦ catalyzes crRNA maturation without a tracrRNA and within the RuvC domain – unlike other well-characterized Cas enzymes (e.g., Cas9 and Cas12a), which use one (Cas12a) or two (Cas9) active sites for DNA cutting and harbor a distinct active site (Cas12a) or additional factors (Cas9) for crRNA processing.
After understanding how CasΦ functions, the researchers discovered it is a functional genome editor by using a GFP disruption assay in HEK293T cells, with CasΦ-2 having comparable editing efficacy to well-described systems such as CRISPR-Cas9. To further confirm gene editing capabilities, CasΦ-2 was delivered as ribonucleoproteins into plant protoplast to successfully edit the A. thaliana PDS3 gene, which primarily produced 8- to 10-bp deletions.
A single active site for crRNA processing and DNA cutting suggests CasΦ proteins may be particularly amenable to engineering, expanding functionality for genetic manipulation. Additionally, minimal PAM requirements and its compact size could be beneficial for vector-based delivery into cells and a more comprehensive range of targetable genomic sequences.
Pausch et al. have characterized a hypercompact Cas protein found in huge phages and have verified that it is a functional genetic editor with a unique mechanism and minimal requirements. This work provides the opportunity to use an efficient and small gene editor and justifies exploring other huge phage enzymes, which could overcome barriers and limitations of current technologies.
Newly characterized and validated CRISPR-Cas systems expand the gene-editing toolbox, providing researchers with new avenues to get their science done and impact wide-ranging fields, from fundamental research to medicine.
References
- Al-Shayeb, Basem, et al. "Clades of huge phages from across Earth's ecosystems." Nature 578.7795 (2020): 425-431.https://doi.org/10.1038/s41586-020-2007-4
- Pausch, Patrick, et al. "CRISPR-CasΦ from huge phages is a hypercompact genome editor." Science 369.6501 (2020): 333-337. DOI: 10.1126/science.abb1400