Wastewater Testing for Infectious Disease Surveillance
Wastewater testing has long been used to uncover concerning pathogens in our water supply. However, with the rise of more complex viral pathogens, the practice has taken on a new layer of value for communities around the world. Next generation sequencing (NGS) has played a significant role in expanding wastewater testing and therefore the adoption of this tool by additional organizations. In recent years, we’ve seen unprecedented use by public health organizations, colleges and universities, as well as environmental surveillance groups. The emergence of NGS has undoubtedly provided these researchers with a powerful tool to identify, track, and trace even the most complex viral challenges, including SARS-CoV-2 and monkeypox.
Yet, given cost constraints, the need for expert bioinformatics analysis, varied physiochemical water characteristics, lack of standard sampling practices, and the continued changing pace of technology, the widespread adoption of wastewater testing has been limited.
To overcome these barriers, research teams in numerous countries have deployed projects aimed at highlighting the benefits associated with using wastewater testing as an early-onset monitoring system to provide public health officials with critical information regarding the spread of infectious viral strains. Here, we’ll take a closer look at how Integraded DNA Technologies (IDT) and their partners at Psomagen® collaborated to overcome these barriers and position NGS as a leading technology for wastewater surveillance. Psomagen is a member of the IDT AlignSM program and led the sequencing and analysis efforts of this study.
Case Study: Identifying emerging variants sooner
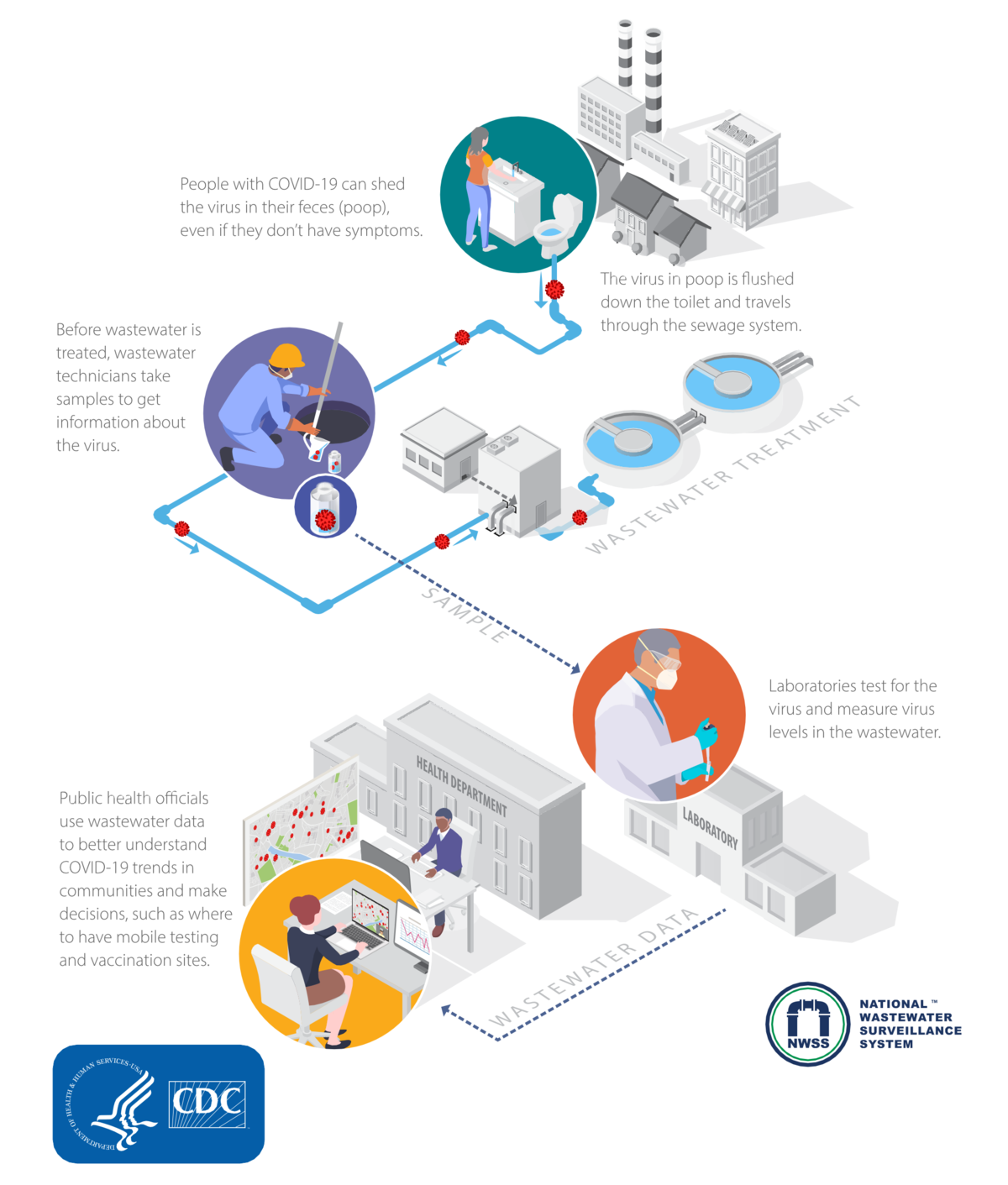
People infected with a virus, such as SARS-CoV-2, can shed this virus in their urine and feces, even if they don’t have symptoms. The virus can then be detected in wastewater, enabling wastewater surveillance to capture presence of certain infectious diseases that are shed by people with and without symptoms. This allows wastewater surveillance to serve as an early warning for emerging viral outbreaks within a community. Once health departments are aware, communities can act quickly to prevent the spread of the virus. Data from wastewater testing support public health mitigation strategies by providing additional crucial information about the prevalence of a particular virus within a community (Diagram 1).
Data from wastewater testing are meant to complement existing viral surveillance systems by providing [1]:
- Greater access to samples: Wastewater-Based Epidemiology (WBE) can provide an efficient and cost-effective method to access samples at greater scale, especially for counties where clinical testing is underused or unavailable.
- More informed decision making: WBE can improve intervention strategies by providing a more accurate assessment of disease progression within a given community.
- Faster results: WBE has been shown to be an effective early warning system for new infectious diseas outbreaks at a community level.
IDT in partnership with Psomagen and Mike Wiley, PhD, Assistant Professor at the University of Nebraska Medical Center set out to determine the most appropriate assay design and chemistry for the surveillance of emerging viral strains in wastewater samples. In this study, the SARS-CoV-2 virus was monitored across various municipalities within the state of Nebraska using two different amplicon sequencing based approaches.
Diagram 1. A visual depiction of how wastewater surveillance is conducted provided by the CDC and National Wastewater Surveillance System
Materials developed by CDC. The use of this material with any reference to IDT products or services does not constitute endorsement or recommendation by the National Wastewater Surveillance System, US Government, Department of Health and Human services, or Centers for Disease Control and Prevention. This diagram is available at How Wastewater Surveillance Works [2].
As highlighted in previous published studies, there are unique challenges associated with wastewater surveillance studies that make sequencing of these samples difficult. Some of these include [3]:
- Insufficient coverage of the entire genome across multiple samples.
- Limited amounts of RNA within each sample with varying concentrations.
- Labor intensive and long turnaround times associated with traditional sequencing approaches.
- Mixture of viral and bacterial genomic material.
Here, we present a novel approach for overcoming common challenges associated with wastewater surveillance for infectious disease outbreaks.
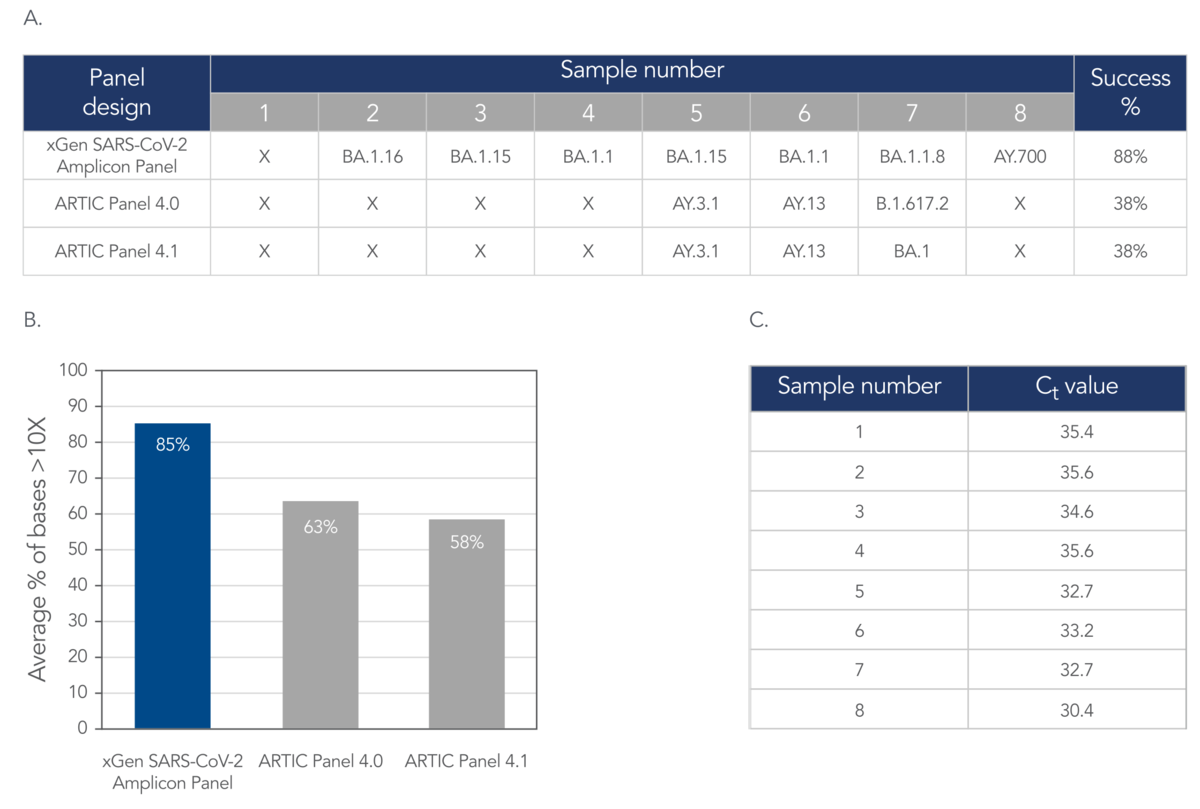
Over the course of the pandemic, several amplicon-based approaches emerged into prominence amongst researchers around the world, such as the ARTIC and Midnight workflows. Though these methods provide a sufficient level of genomic coverage and depth of sequencing for most viral surveillance approaches, we sought an alternative amplicon-based workflow that is better suited to overcome common challenges associated with WBE, such as sample-to-sample variances due to either low input amounts or high Ct values. We compared our novel amplicon chemistry against two ARTIC primer design sets across multiple wastewater samples. As shown in Figure 2, the xGen SARS-CoV-2 Amplicon Panel achieved not only a higher average of total bases covered but also produced less sample-to-sample variance.
Figure 2. The xGen SARS-CoV-2 panel obtainted better pango lineage confirmations and percentage of bases covered above 10X when compared to the two ARTIC panel designs. (A) Lineage confirmation results from all three panel designs (B) Average percent of % bases greater than 10% (C) Ct values associated with each sample (A-C) Total RNA was was prepared from wastewater samples (n = 8) that were known to be infected with the SARS-CoV-2 virus. The resulting NGS libraries generated with the xGen SARS-CoV-2 Amplicon Panel were sequenced 2 x 150 bp PE on a NovaSeq® System (Illumina). The resulting NGS liibraries generated with the SARS-CoV-2 ARTIC Amplicon Panels were sequenced 2 x 250 bp PE on a MiSeq® System (Illumina). Lineages were called with Pangolin.
The power of super amplicons
We believe the improved results presented in Figure 2, are due to the novel primer design of the xGen SARS-CoV-2 Amplicon Panel.
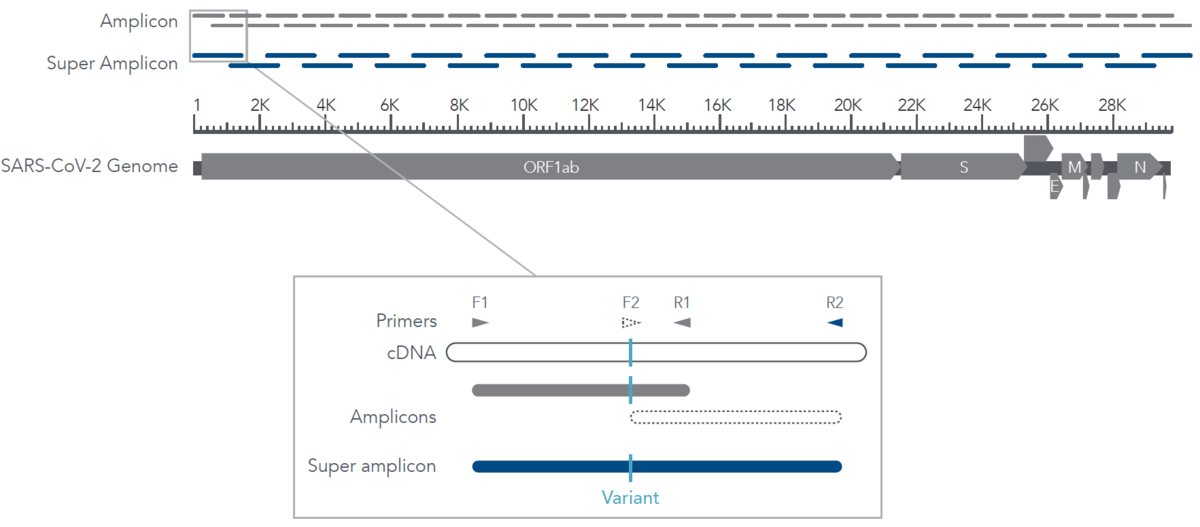
IDT’s one-tube, overlapping primers provide built-in redundancy for novel variant discovery. The creation of “super amplicons” maintains genomic coverage, even in the presence of mutations that result in primer dropout. This unique feature is found in a single-tube workflow since super amplicons do not form in a two-tube workflow, commonly found in traditional amplicon based approaches such as the ARTIC and Midnight primer sets.
A novel variant that causes a loss in primer function could result in lost coverage and missed mutations. In the SARS‑CoV‑2 pandemic, new mutations are regularly being discovered and variant tracking can give important insights. A mutation that occurs in a primer binding site (as represented by a “|” in Figure 3) can cause a loss of primer function (F2). Without the primer binding, the amplicon (white dashed segment) is not created, and the subsequent sequence data for this region is not generated. Luckily, both a one-tube and a two-tube technology will be able to observe this mutation using the adjacent amplicon (gray). In a one-tube solution, primers F1 and R2 are in close enough proximity to create a super amplicon (blue) thereby allowing one to still observe the mutation while also avoiding any loss in total genomic coverage. Traditional amplicon methods lack the formation of these super amplicons, thereby increasing the chances of primer dropouts which could lead to novel mutations being missed.
Figure 3. Details of the formation of super amplicons across the SARS-CoV-2 genome. Proper identification is crucial for tracking nucleotide variants and understanding virus evolution, transmission, and pathogenesis. The IDT xGen Amplicon Technology increases your confidence in the sequence data obtained from novel variants. Since primer sequences do not need to be redesigned, the IDT xGen Amplicon Technology reduces the time and resources needed to revalidate new versions of amplicon panels found in other methods.
Conclusion: Benefits of wastewater surveillance
This study underscores the potential value found in using NGS technologies to inform, guide, and uncover essential data as we seek to better understand evolving epidemiological patterns. In conclusion, several benefits can be realized when using NGS for wastewater surveillance, including:
- Early warning system: In cases where initial diagnostic testing is unavailable or requires lengthy processing intervals, wastewater testing offers health officials close to real-time insights on disease prevalence. This benefit is critically important in situations where novel viruses are at play, as diagnostic testing development takes time. For example, in the SARS-CoV-2 outbreak of 2020, rapid forms of diagnostic testing took months to develop, manufacture, and distribute. During this time, wastewater testing filled a critical information gap.
- Communal transmission surveillance: When days, if not hours, matter, this large-scale effort reduces cost and effort, while also saving significant time.
- Cost effective: As the use of NGS technologies continues, wastewater surveillance will enable research teams to efficiently survey hundreds to thousands of individuals at one time, which is significantly more cost effective than individual diagnostic tests.
- Reduced bias: Although unintentional, when working on a global scale, socioeconomic, geographic, and personal biases can enter the statistical equation. In the case of COVID-19, these biases were especially problematic given that initial disease prevalence monitoring was based on the number of positive counts, yet community access to testing was extremely limited.
IDT - your partner for Next Generation Sequencing
Integrated DNA Technologies (IDT) is your advocate for the genomics age. They produce tools for NGS, CRISPR, qPCR and PCR. Their products include DNA/RNA oligos, genes and gene fragments. For more than 30 years, IDT's innovative tools and solutions for genomics applications have been driving advances that inspire scientists to dream big and achieve their next breakthroughs.

xGen™ NGS Amplicon Sequencing
Our amplicon solutions enable you to advance from sample to sequencing faster, without sacrificing coverage or yields. If your research requires rapid, ready-to-sequence materials or you are working with low-input quantities, xGen NGS can unlock the answers you seek.
xGen™ SARS-CoV-2 Amplicon Panels
Targeted sequencing to research SARS-CoV-2 viral genomics
Overlapping primers produce 99.7%† genomic coverage of SARS-CoV-2 lineages. These consistent primers obtain sequence data from viral concentrations as low as 10–100 viral copies without the need to evaluate new primer sets for emerging lineages.
References
- Ai Y, Davis A, Jones D, et al. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci Total Environ. 2021;801:149757.
- How wastewater surveillance works. 2022; www.cdc.gov/healthywater/surveillance/wastewatersurveillance/resources/how-wws-works.html. Accessed October 14, 2022.
- Karthikeyan S, Levy JI, De Hoff P, et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature. 2022;609(7925):101-108.