The problem
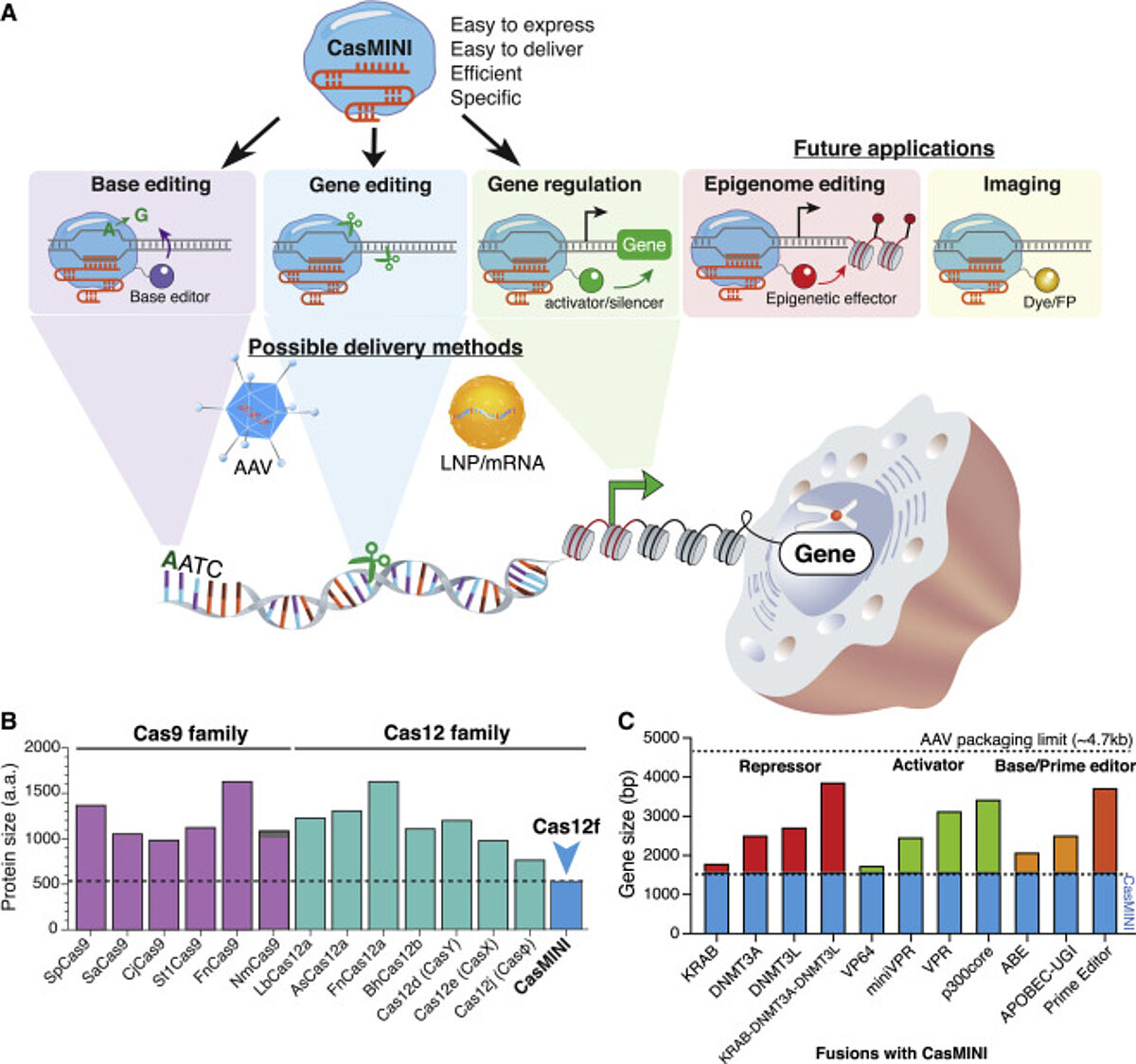
CRISPR-Cas systems have revolutionized genome engineering, offering opportunities for the development of gene therapies against a variety of human diseases. However, their large sizes often restrict delivery into cells, impeding clinical utility. For instance, Cas9 and Cas12a—nucleases capable of specific genome editing—are typically 1,000-1,500 amino acids, which doesn’t leave much room for guide RNAs (gRNA) and regulatory elements when considering the packaging capacity of adeno-associated viral vectors (<4.7 kb).
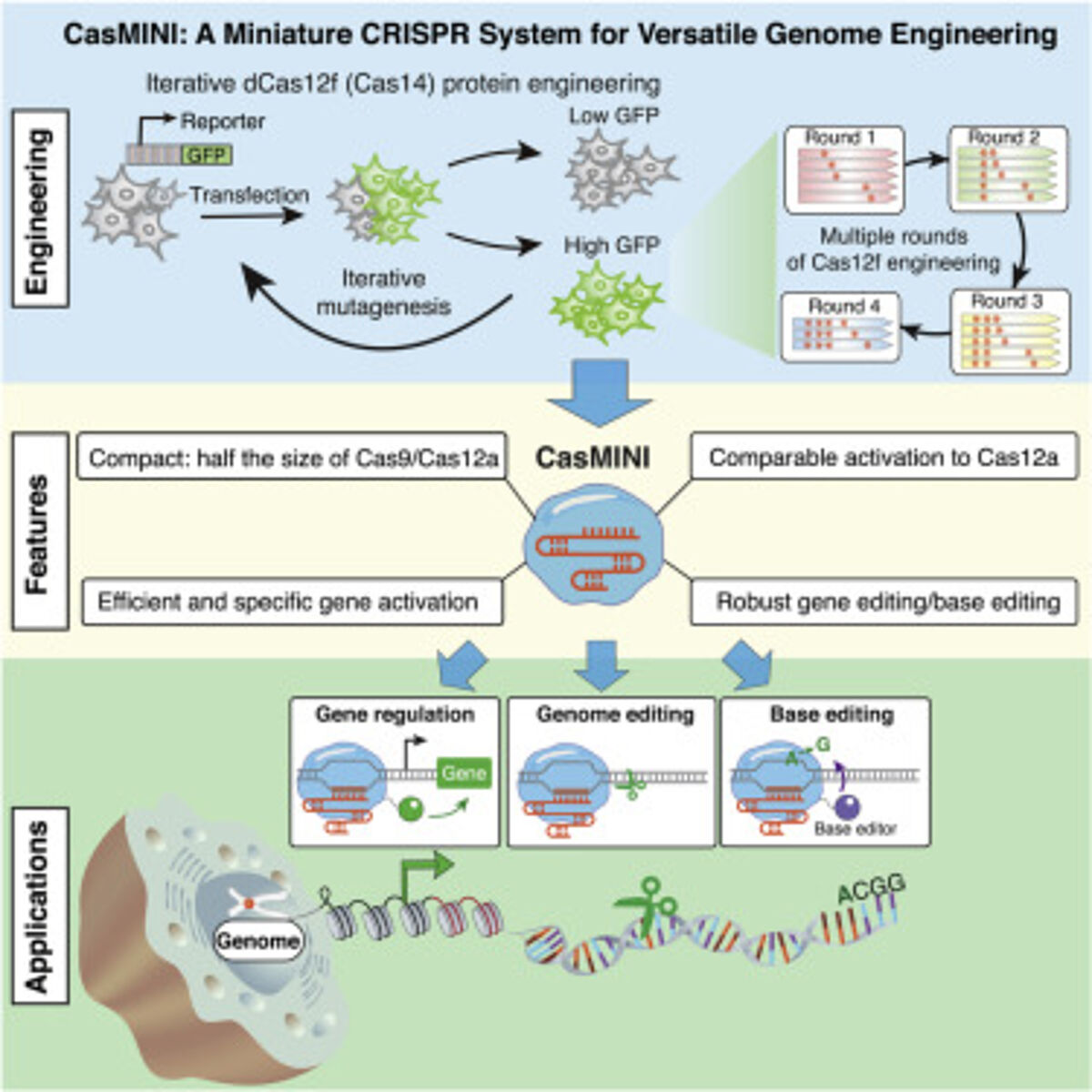
So, how can we get CRISPR-Cas systems into cells to do their job? Make them smaller. This is easier said than done. While smaller naturally occurring Cas effectors exist (e.g., Cas12f and Cas12j), they are not necessarily functional in mammalian cells. Xu et al., explore smaller Cas proteins to develop more compact—and functional—CRISPR-Cas systems and, ultimately, overcome cell delivery challenges.
The solution
To enhance the toolkit for gene editing and base editing, the researchers applied RNA engineering and protein engineering to type V-F Cas12f—a 529 amino-acid nuclease with no detectable activity in mammalian cells (also revealed through this work).
Their goal was to generate a compact, efficient, and specific system for mammalian genome engineering. They were successful and dubbed the new system CasMINI.
A little information about Cas12f
CRISPR-Cas12f, is a family of exceptionally compact RNA-guided nucleases from uncultivated archaea. Known to cut single-stranded DNA (ssDNA), the wild-type Cas12f system also possesses protospacer adjacent motif (PAM) sequences for double-stranded DNA cleavage in vitro.
The approach
By optimizing the single guide RNA (sgRNA) design and performing multiple rounds of iterative protein engineering and screening Xu et al., generated Cas12f variants to obtain:
- Nuclease-deactivated CasMINI (dCasMINI) that, when fused with a transcriptional activator, could activate reporter and endogenous gene expression;
- Nuclease-deactivated CasMINI with base editing capabilities; and
- Nuclease-active CasMINI with efficient and distinct genome editing capabilities.
The findings
Xu et. al developed a miniature CRISPR system using Cas12f for genome engineering that functions robustly in mammalian cells. The engineered CasMINI is compact and less than half the size of Cas9 and Cas12a.
Gene Activation
The researchers generated a dCasMINI with significant improvement in gene activation over the wild-type dCas12f system and comparable activation ability with the dCas12a system (see the table below for specific mutations and their impact). Through whole-transcriptome RNA sequencing, the researchers found that dCasMINI is specific in mammalian cells (comparable with dCas12a).
| Construct | Mutation | Performance |
|---|---|---|
| dCas12f | D326A and D510A | Deactivated nuclease activity in Cas12f (all dCasMINI constructs have these mutations) |
| dCasMINI-V1 | D143R | 123-fold gene activation compared with a non-targeting sgRNA, which was more than 34-fold improvement over the wild-type dCas12f |
| dCasMINI-V2 | D143R/T147R | 1.55-fold improvement in activation over dCasMINI-V1 |
| dCasMINI-V3 | D143R/T147R/K330R | 1.26-fold improvement over the best double variant |
| dCasMINI | D143R/T147R/K330R/E528R | 1.14-fold improvement over the best triple variant and almost 200-fold improvement of reporter gene activation over the wild-type Cas12f |
All of these mutations reside in the DNA binding pocket.
With an optimized sgRNA, dCasMINI efficiently activated genes endogenous to HEK293T cells including HBG, IL1RN, ASCL1, IFN????, CD2, CXCR4, and HBB. What’s more, dCasMINI outperformed dCas12f for endogenous gene activation and is comparable with dCas12a. For example, IFNg activation with dCasMINI reached a 300-fold improvement over dCas12f on the mRNA level and 768-fold on the protein level when co-delivering two sgRNAs.
Base Editing
By fusing dCasMINI to the deoxyadenosine deaminase domain heterodimer (TadA-TadA-8e), the researchers observed detectable A-T to G-C base conversion at several genomic sites by using high-throughput sequencing. Base editing efficiency was dependent on the target site and was most efficient 3-4 bp downstream of the PAM.
Gene Editing
Using nuclease-active versions of dCasMINI variants (CasMINI), the researchers observed
robust gene editing. Specifically, adding an additional mutation (E151A) to CasMINI-V3 resulted in more efficient indel formation at four genomic sites in HBB and IFN???? compared to wild-type Cas12f and CasMINI-V2. Additionally, nuclease-active CasMINI resulted in distinct editing patterns from Cas9.
The impact
CasMINI has great potential for broad genome engineering applications where compact Cas fusion proteins for delivery and cellular function are required. This work suggests that many Cas12 proteins could be optimized for better efficiency and functionality in mammalian cells through protein and guide RNA engineering. The RNA and protein engineering approach used by Xu et al., could be useful in engineering more Cas12f/Cas14 effectors from other bacterial or archaeal species to expand the genome engineering toolkit and overcome cell delivery challenges.
Read the original article
Xu, Xiaoshu, et al. "Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing." Molecular Cell (2021). https://doi.org/10.1016/j.molcel.2021.08.008