As of June 2021, over 172 million people have been diagnosed with COVID-19 and over 3.7 million have died from the virus globally (https://coronavirus.jhu.edu/map.html). Fortunately, over 2 billion people worldwide have been vaccinated for COVID-19, however, infection and future outbreaks continue to threaten the world. The development of broad-spectrum anti-coronavirus drugs could help address the current crisis, and potentially quickly contain outbreaks in the future.
Our options to prevent or treat coronavirus infections remain limited. To develop broad-spectrum antivirals, it is crucial to understand which host factors coronaviruses require to infect a cell, however beyond the entry step of viral infection, host-virus interplay throughout the viral life cycle remains unknown.
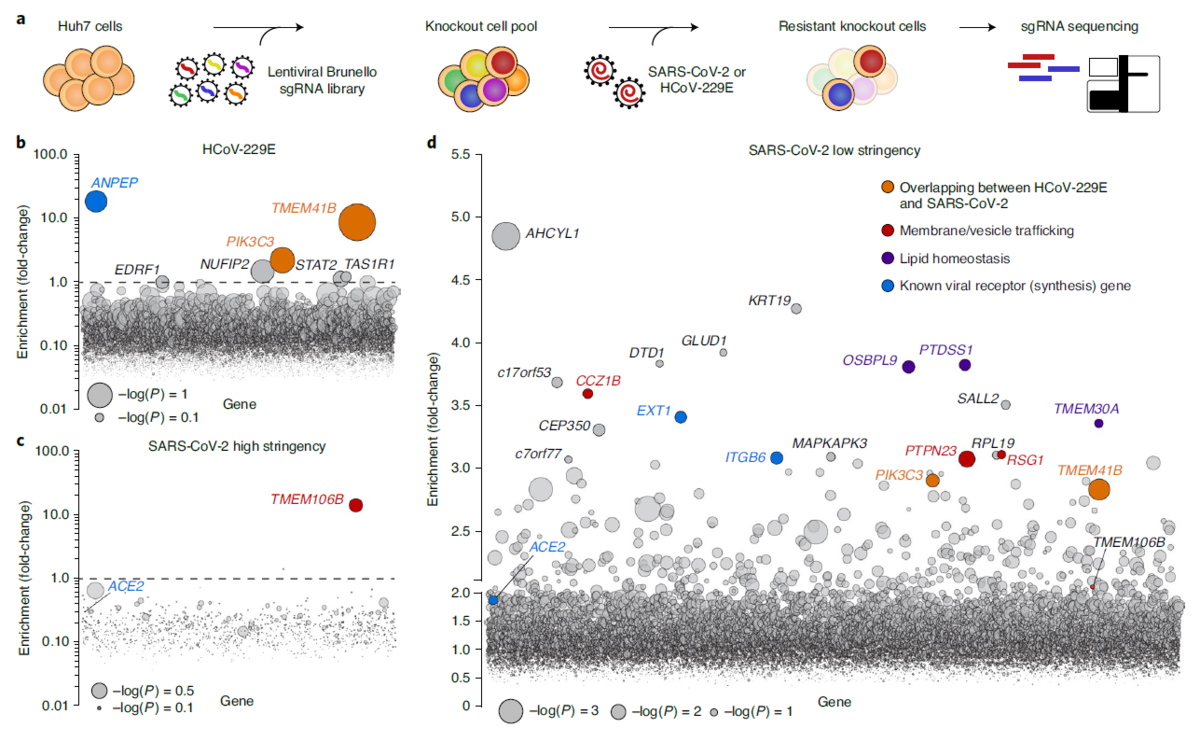
Baggen et. al., have now identified host factor requirements for coronavirus infection by using a series of genome-wide CRISPR-based genetic screens, validating them in various assays, and comparing the results to other screening methods.
The approach
Baggen et. al., performed a high-stringency and lower-stringency CRISPR-based genome-wide screen for SARS-CoV-2 and for the less pathogenic HCoV-229E and HCoV-OC43 to identify both broad-spectrum coronavirus host factors and specific host factors for each virus.
After validating the top hits for SARS-CoV-2, the researchers performed knockout studies and single-cell RNA-sequencing of samples from patients that were COVID-19 positive to further confirm infection requirements and get an idea of how gene expression relates to infection.
The findings
The researchers identified a common host factor required for infection with SARS-CoV-2, HCoV-229, and HCoV-OC43, and also found crucial host factors for HCoV-229E and SARS-CoV-2.
Phosphoinositide 3-kinase (PI3K) type 3 as a common host factor for coronaviruses
Through CRISPR-based screening, Baggen et. al., identified phosphoinositide 3-kinase (PI3K) type 3 as a common host factor for SARS-CoV-2, HCoV-229E and HCoV-OC43. PI3K plays a role in many processes, such as endocytic trafficking and the initiation and maturation of autophagosomes. Inhibiting this protein might serve as broad approach for treating coronavirus, however it is important to note that the researchers did find that coronaviruses employ PI3K for infection but do not depend on a functional macroautophagy pathway.
TMEM41B is a crucial host factor for HCoV-229E infection
TMEM41B was identified as a host factor in the HCoV-229E CRISPR-based high-stringency screen, and TMEM41B knockout in Huh7 cells almost completely protected against infection—making this autophagy protein indispensable for HCoV-229 infection.
TMEM106B is required for SARS-CoV-2 infection
The high-stringency SARS-CoV-2 screen identified TMEM106B—a major player for regulating lysosomal size, motility and responsiveness to stress—as a required host factor for SARS-CoV-2 infection. Several liver and lung-derived cell lines without TMEM106B (via knockout) resisted SARS-CoV-2 infection and reduced virus replication—making this transmembrane protein a crucial proviral host factor for productive SARS-CoV-2 infection.
TMEM106B expression correlates with SARS-CoV-2 infection
Using single-cell RNA-sequencing (scRNA-seq), the researchers were able to analyze TMEM106B expression in bronchoalveolar lavages from 44 patients with pneumonia—where 31 patients were COVID-19 positive. After analyzing 116,797 cells, TMEM106B expression was significantly higher in epithelial cells from patients with COVID-19 compared with epithelial cells from uninfected controls.
Comparison to Other Studies
Baggen et. al., are not the only ones using genome-wide CRISPR screens to identify host factors for SARS-CoV-2, and they used this to their advantage to compare screening results. Overall, they found that there was limited overlap between screens, which may be due to technical variations, such as the CRISPR library, multiplicity of infection (MOI), virus strain or cell line use. In particular, screens performed in similar cell types demonstrated strong overlap
Nonetheless, top-ranked genes that Baggen et. al., identified for HCoV-229E screen (ANPEP, PIK3C3 and TMEM41B) were identified in the other screens, and TMEM106B was found in three other screens.
A little introduction to TMEM106B
TMEM106B is a poorly characterized transmembrane protein within endosomes and lysosomes that is known to control lysosome size, number, mobility and trafficking, as well as play a pivotal role in lysosomal acidification. Recently, it has been noticed for its role in dementia—and now thanks to Baggen et. al., and other teams, for its role in SARS-CoV-2 infection.
The impact
As the world has witnessed over the course of the past 1.5 years, devastating pathogens can emerge and create global health and economic crises. Situations like these will periodically arise beyond the current pandemic, and we need to be better prepared—Baggen et. a., along with numerous other research teams, are working diligently to turn preparedness into a reality. The essential coronavirus host factors identified in these CRISPR-based genome-wide screens will help guide future studies for elucidating the pathways coronaviruses use to hijack our cells, and can potentially serve as targets for treating and preventing COVID-19 or future coronaviruses infections.
References
Baggen, J., Persoons, L., Vanstreels, E. et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat Genet 53, 435–444 (2021). https://doi.org/10.1038/s41588-021-00805-2
Visit our COVID-19 reagents page