CRISPR - which works better, sgRNAs or two-part cr/tracr RNAs?
The CRISPR-Cas9 system has accelerated biomedical research by allowing straightforward and efficient editing of the genome at specific locations. Editing is achieved by generation of double-stranded breaks that either can lead to insertions or deletions and subsequent knockout of genes via the non-homologous end-joining (NHEJ) pathway or can lead to activation of the homology-directed repair (HDR) process when a template is present. HDR can be used to correct mutations or to introduce novel genetic sequences. For instance, HDR could be used to produce fusion proteins.
CRISPR-Cas9 editing: 2 guide RNA options
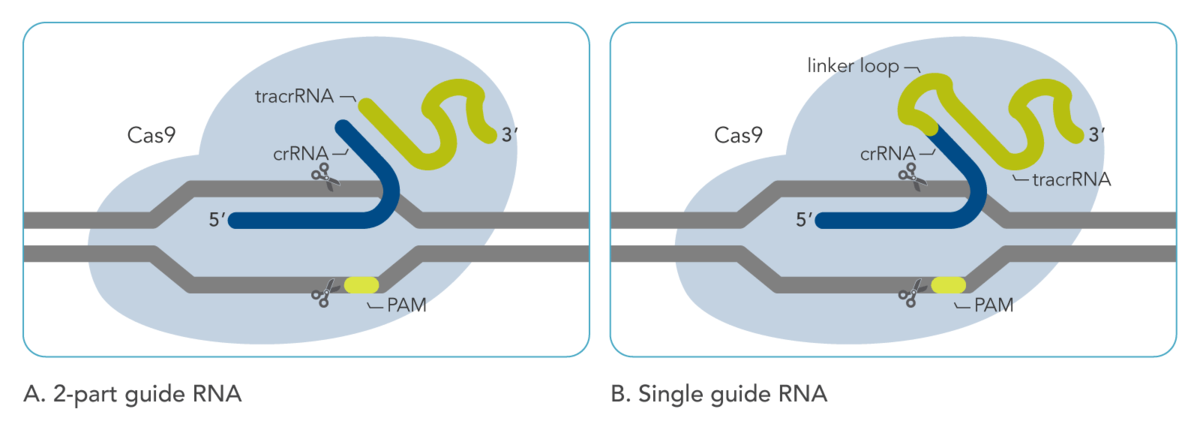
During CRISPR-Cas9 editing, double-stranded breaks in genomic DNA are generated by the Cas9 protein. The specificity of DNA cleavage depends on a guide RNA molecule that is necessary for the Cas9 nuclease to be active. This guide RNA consists of a constant region (which binds Cas9) and a variable region, called the spacer, which directs the Cas9 nuclease to a homologous target protospacer location in the genome. In native bacterial immune systems, the Cas9 guide RNA is not just one molecule; it is a duplex made of a tracrRNA and a crRNA (Figure 1A). These two RNA molecules hybridize together by a 16-nt complementary sequence. The constant region is present on the tracrRNA, while the spacer is present on the crRNA molecule.
Which is better, 2-part or single guide RNA?
Some reports have stated that single guide RNAs outperform 2-part guide RNAs in most or all cases. Here, we compare the 2 types of guide RNA complexes directly. First, 255 target sites were randomly selected across the genome. RNP complexes were made using either 2-part or single guide RNA and delivered into Jurkat cells. Genome editing efficiencies were determined. Single guide RNA editing efficiencies for each target site were plotted against the 2-part guide RNA editing efficiencies. The majority of target sites (189 out of 255 target sites, 74%) showed genome editing levels of >80%, independent of guide RNA system. Explore detailed results in the original article...