Ligases, DNA polymerases, and RNA polymerases molecular biology enzymes with distrinct functions and applications:
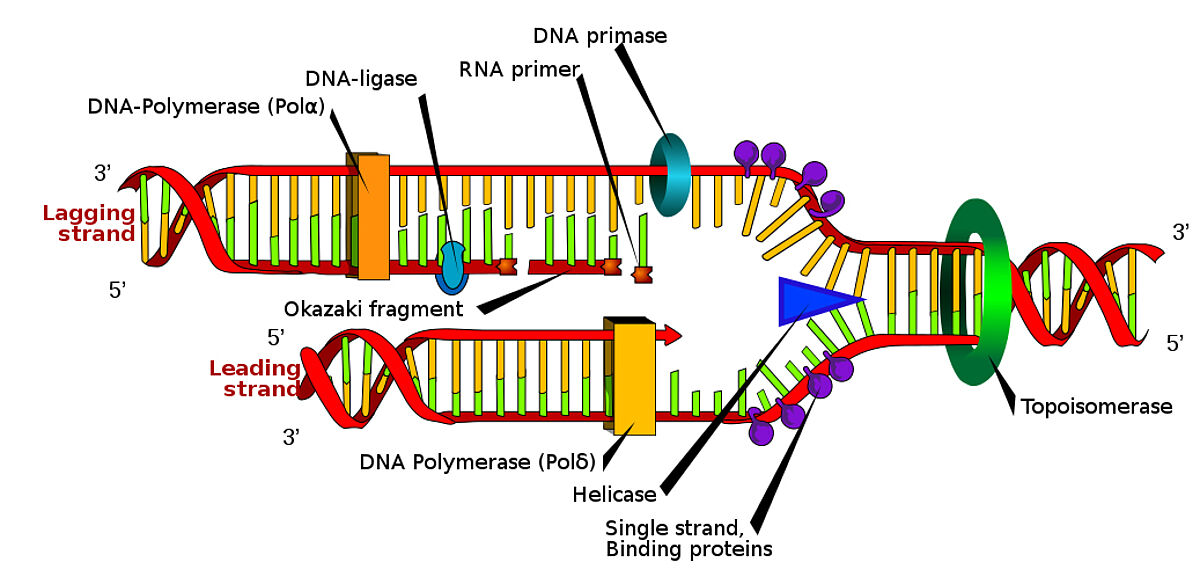
Ligases: Ligases are enzymes that catalyze the formation of phosphodiester bonds between DNA or RNA molecules. They are responsible for joining fragmented DNA strands, repairing DNA damage, and connecting DNA or RNA molecules. Ligases play a crucial role in sealing the nicks or gaps in the sugar-phosphate backbone of DNA or RNA, ensuring the continuity and integrity of genetic material.
DNA Polymerases: DNA polymerases are enzymes that catalyze the synthesis of new DNA strands by adding complementary nucleotides to a DNA template strand during DNA replication and repair. They work in a highly accurate and processive manner, ensuring faithful duplication of DNA. DNA polymerases also have proofreading mechanisms to correct errors that may occur during DNA synthesis, contributing to their crucial role in maintaining genomic stability.
RNA Polymerases: RNA polymerases are enzymes responsible for transcribing DNA sequences into complementary RNA strands. During transcription, RNA polymerases recognize specific DNA sequences known as promoters and initiate the synthesis of RNA molecules from a DNA template. There are different types of RNA polymerases in eukaryotes and prokaryotes, each responsible for synthesizing different classes of RNA, such as mRNA (messenger RNA), rRNA (ribosomal RNA), and tRNA (transfer RNA). RNA polymerases play a vital role in gene expression, as they are involved in producing the templates for protein synthesis.
Ligase
NxGen T4 DNA Ligase
NxGen T4 DNA Ligase is a ATP-dependent ligase commonly used for DNA cloning. It covalently joins dsDNA molecules having 5′-phosphorylated and 3′-hydroxylated blunt or compatible cohesive ends produced by restriction enzyme digestion or other enzymatic processes. It has no activity on single-stranded nucleic acids.
- Ligation of blunt or cohesive-ended DNA fragments
- Repair of nicks in double-stranded nucleic acids
Ampligase Thermostable DNA Ligase
Derived from a thermophilic bacterium, Ampligase Thermostable DNA Ligase is stable and active at much higher temperatures than conventional DNA ligases. This enzyme catalyses NAD-dependent ligation of adjacent 3′-hydroxylated and 5′-phosphorylated termini in duplex DNA structures that are stable at high temperatures. Its half-life is 48 hours at 65 °C and >1 hour at 95 °C. It is active for at least 500 thermal cycles (94 °C/80 °C) or 16 hours of cycling, which permits extremely high hybridisation stringency and ligation specificity. It has no detectable activity in ligating blunt-ended DNA, RNA, or RNA:DNA hybrids.
- Sensitive detection of single-nucleotide polymorphisms (SNPs)
- Ligation amplification (ligase chain reaction, LCR)
- Gibson Assembly cloning
- Repeat expansion detection (RED)
- Simultaneous mutagenesis of multiple sites
T4 RNA Ligase 2, Deletion Mutant
T4 RNA Ligase 2, Deletion Mutant, T4Rnl2(1-249), ligates single-stranded, adenylated DNA or RNA (App-DNA or App-RNA) oligonucleotides to small RNAs. The preadenylated 5′ ends of DNA or RNA are ligated to the 3′ ends of RNA in the absence of ATP, which prevents circularisation and other undesirable bimolecular reactions.
- Preparation of cDNA libraries for small-RNA transcriptome analysis, such as RNA-Seq
- Optimal linker ligation for miRNA cloning
CircLigase II ssDNA Ligase
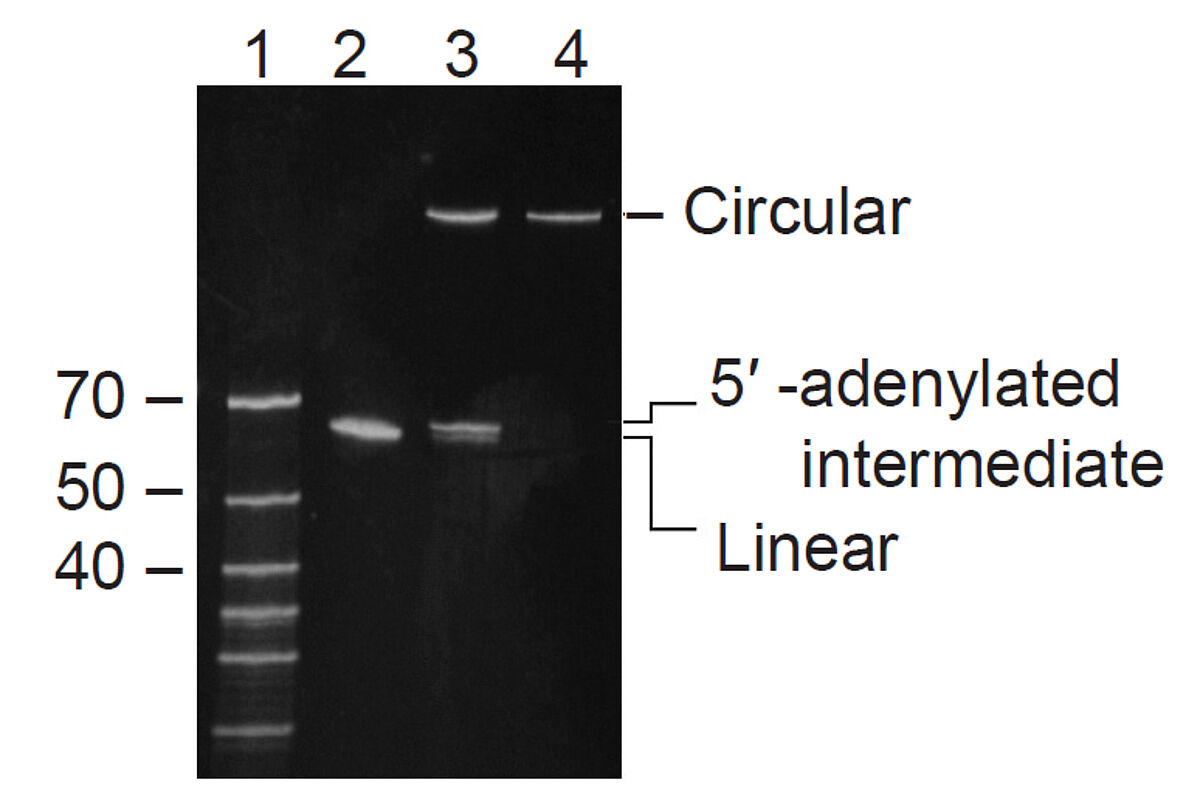
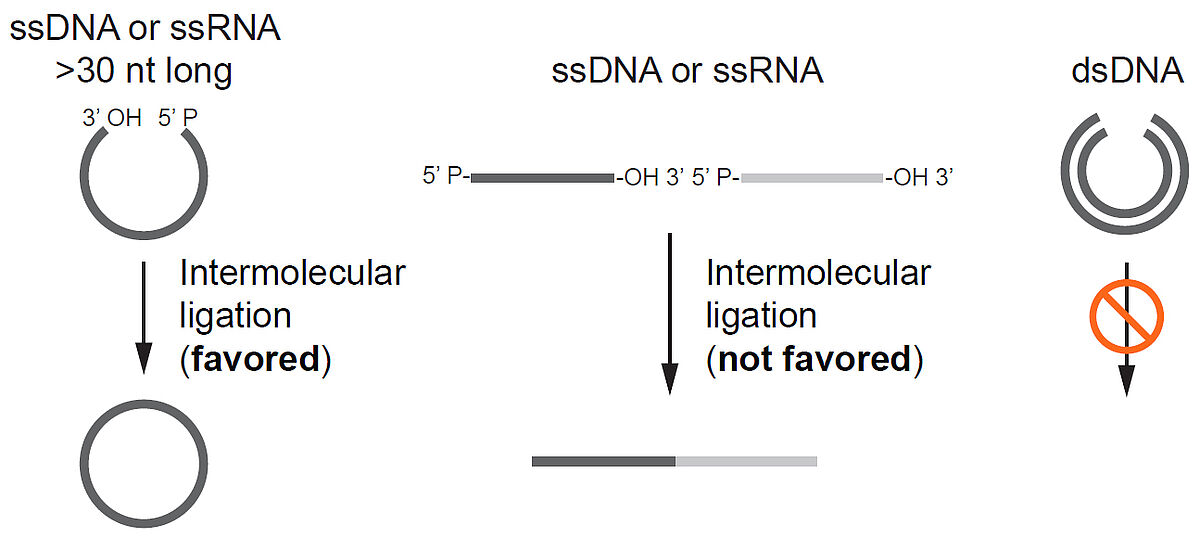
This thermostable ligase catalyses intramolecular ligation (i.e., circularisation) of ssDNA templates >30 nt having a 5′ phosphate and a 3′ hydroxyl group. It ligates the ends of ssDNA in the absence of a complementary sequence. Standard reaction conditions produce no detectable single-stranded DNA concatamers or concatameric DNA circles. Due to the high degree of adenylation, a single CircLigase II enzyme can ligate only a single molecule of nonadenylated DNA, and the reaction stops in the absence of ATP. Therefore, a 1:1 stoichiometric amount of ligase:substrate is required to drive the ligation reaction to completion.
- Production of ssDNA templates for rolling-circle replication or rolling-circle transcription experiments
- Production of ssDNA templates for RNA polymerase and RNA polymerase inhibitor assays
CircLigase ssDNA Ligase
This thermostable, ATP-dependent ligase catalyses intramolecular ligation (i.e., circularisation) of ssDNA templates >30 nt having a 5′ phosphate and a 3′ hydroxyl group. It ligates the ends of ssDNA in the absence of a complementary sequence. Standard reaction conditions produce no detectable single-stranded DNA concatamers or concatameric DNA circles. Due to the low degree of adenylation, CircLigase enzyme has high turnover; it can reversibly and repeatedly act on multiple preadenylated DNA molecules under nonstoichiometric reaction conditions.
- Production of ssDNA templates for rolling-circle replication or rolling-circle transcription experiments
- Production of ssDNA templates for RNA polymerase and RNA polymerase inhibitor assays