Guidelines for achieving high rates of HDR with CRISPR Cas9
Double-strand breaks (DSBs) in genomic DNA are repaired by a variety of pathways [1]. The non-homologous end-joining (NHEJ) pathway and similar pathways cause imprecise repair of the DNA sequence, generating random indels. The HDR pathway, however, allows introduction of predictable, precise mutations. HDR depends on the presence of a DNA template with homology to the genomic sequence adjacent to the DSB. In a research setting, repair through NHEJ is sufficient for gene disruption (knockout), but HDR is essential for applications requiring precise repair such as generation of knockin cell lines or model organisms, as well as gene correction by generation of specific point mutations.
Comparison of CRISPR-Cas9 and CRISPR-Cas12a
There are many ways to generate targeted DSBs, and CRISPR-Cas9 is a well-studied system with reliable results. However, Cas12a (also known as Cpf1), another class II CRISPR endonuclease [2], has the potential to overcome some Cas9 limitations, such as Cas9’s G-rich PAM requirement [3]. Cas12a expands the range of potential CRISPR target sites, as it recognizes a T-rich PAM, TTTV (V = A/G/C). Cas12a also displays intrinsically low off-target cleavage [4].
Recent literature has suggested that Cas12a has indiscriminate single-stranded DNA (ssDNA) cleavage activity, making Cas12a incompatible with experiments that use ssDNA donors for HDR. However, although we empirically confirmed this ssDNA cleavage activity, we also found Cas12a to be a fantastic choice for HDR when following simple guidelines. Additionally, one very important improvement we have developed is a novel enzyme variant, Alt-R A.s. Cas12aUltra. By choosing Cas12a Ultra, researchers greatly expand both the number of target sites available for editing and their options for achieving high rates of HDR. In addition, Cas12a Ultra is active at lower temperatures, enabling genome editing of organisms such as fish and plants. With the novel enzyme improvements of Cas12a Ultra and the new HDR guidelines listed below, Cas12a Ultra can be a go-to tool for any genome editing endeavor.
Use a Cas12a nuclease with high cleavage activity, but use at an appropriate concentration
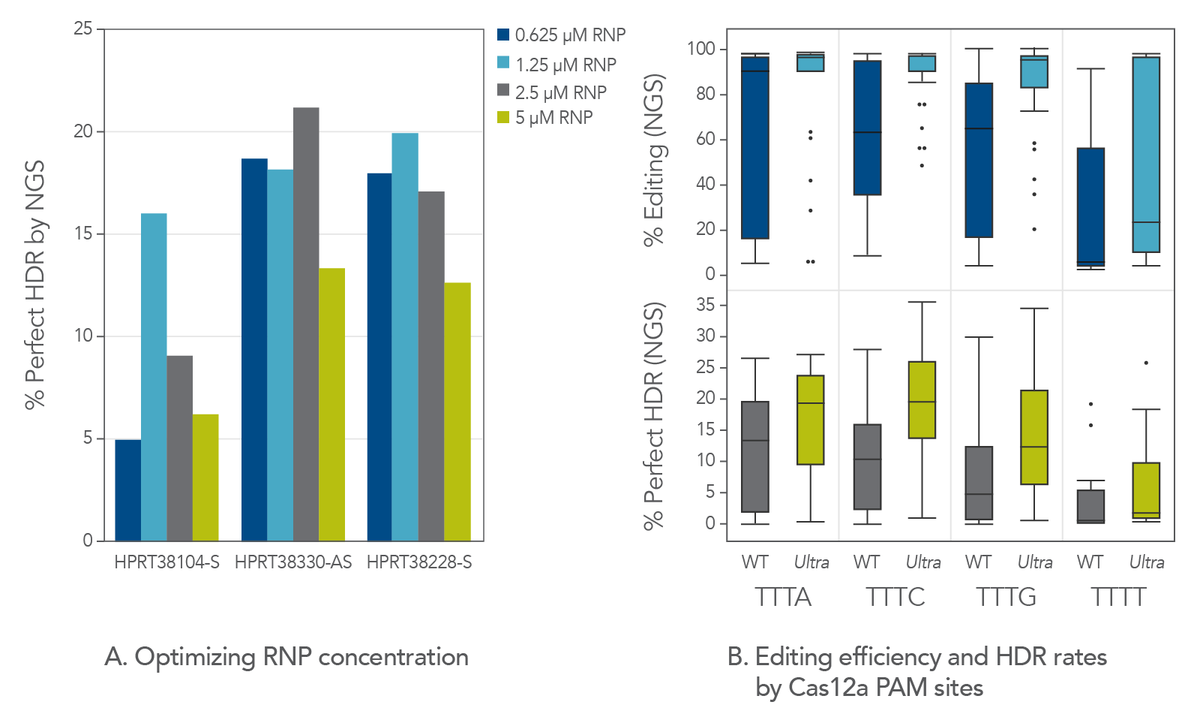
Through directed evolution, IDT has recently developed a Cas12a mutant, Alt-R A.s. Cas12a Ultra, with an improved cleavage rate. Maximizing editing efficiency is essential for maximizing HDR. Cas12a Ultra is an ideal nuclease for mediating both high editing and high HDR rates. However, we observed that too much of this nuclease (e.g., 5 µM) is unnecessary for robust editing and can even be detrimental to HDR. This is likely due to the known ssDNA cleavage activity of Cas12a (Figure 1A). But when Cas12a Ultra is used at an appropriate concentration (1–2 µM RNP), it displays high levels of editing efficiency, and it also increases HDR rates. At these concentrations of RNP, the risk of ssDNA donor degradation is minimized with all Cas12a PAMs, including TTTT (Figure 1B). Optimizing Cas12a Ultra concentration improves editing efficiency and HDR rates at all Cas12a PAMs, even TTTT.
Figure 1. Hyperactive Cas12a Ultra should be used at a moderate concentration for HDR. (A) Jurkat cells were transfected by electroporation with A.s. Cas12a Ultra RNPs targeting the human HPRT genomic locus along with Ultramer HDR donor oligos containing a 6-base insert and 40-nucleotide homology arms. Perfect HDR was measured by NGS. This indicated that HDR is highest when RNP concentration is between 1–2 µM. (B) We designed 110 gRNAs targeting varied genomic regions. Electroporation was performed using RNPs containing either WT A.s. Cas12a or A.s. Cas12a Ultra. Total editing and perfect HDR were measured by NGS. Cas12a Ultra concentration optimization greatly improves editing efficiency, thereby increasing the rate of HDR at all Cas12a
Always use the non-targeted strand as the donor oligo sequence
Cas12a is directed to bind the desired DNA sequence by a 21-nucleotide guide sequence. This is similar to the situation with Cas9, which uses a 19- or 20-nucleotide guide sequence. After binding the target site in the genome, the nuclease domain within Cas12a cleaves both the targeted (guide-complementary; Figure 2, top) and non-targeted strands of the DNA to create a staggered DSB. For mediating repair through HDR, a DNA template with homology to the nucleotide sequence near the DSB is required for precise repair at the desired DNA locus.
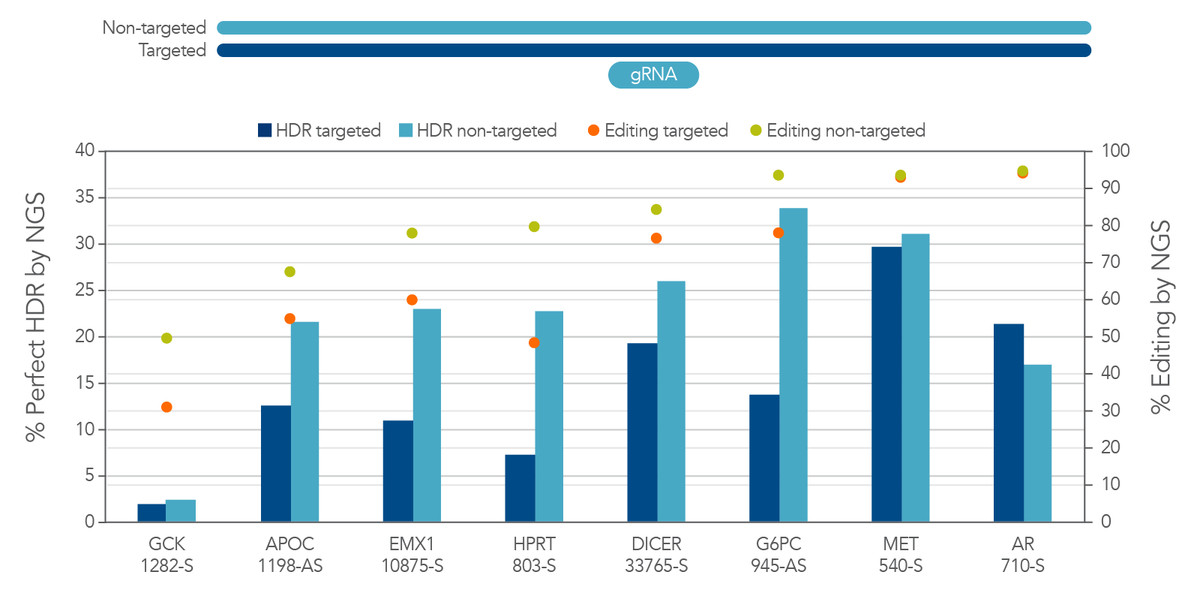
We considered the possibilities of designing this template to contain homology arms containing the sequence from either the targeted or the non-targeted strand. We asked which strand gives better results. To determine the answer to this question, we performed a direct comparison of HDR using targeted and non-targeted ssDNA templates. We discovered that there is strong preference for the non-targeted sequence as the HDR donor (Figure 2B). There is actually a reduction in total editing when a targeted-strand donor is present. This reduction is likely due to interference with Cas12a binding to the desired DNA sequence. This seems to be specific to Cas12a, as this strand preference is not always observed with Cas9. Because of these findings, we recommend researchers always use the non-targeted strand sequence in the homology arms of the donor oligo sequence when using Cas12a.
Figure 2. There is strong preference for using the non-targeted strand sequence as the HDR donor sequence for Cas12a HDR. Jurkat cells were transfected by electroporation with Alt-R A.s. Cas12a Ultra RNPs targeting multiple loci along with Ultramer HDR donor oligos containing a 6-base EcoRI insert and 40-nucleotide homology arms. These single-stranded oligodeoxynucleotides (ssODNs) contained either the targeted or non-targeted strand sequence and were directly compared with each other. Editing efficiency and HDR were measured by NGS. The targeted strand donors had reduced total editing and overall HDR rates, most likely due to interference with Cas12a binding to the desired DNA sequence. In sites where editing was equal, the strong preference for non-targeted sequence was not observed. We recommend always using the non-targeted strand sequence as the donor oligo sequence for HDR when using Cas12a.
Position your insert carefully
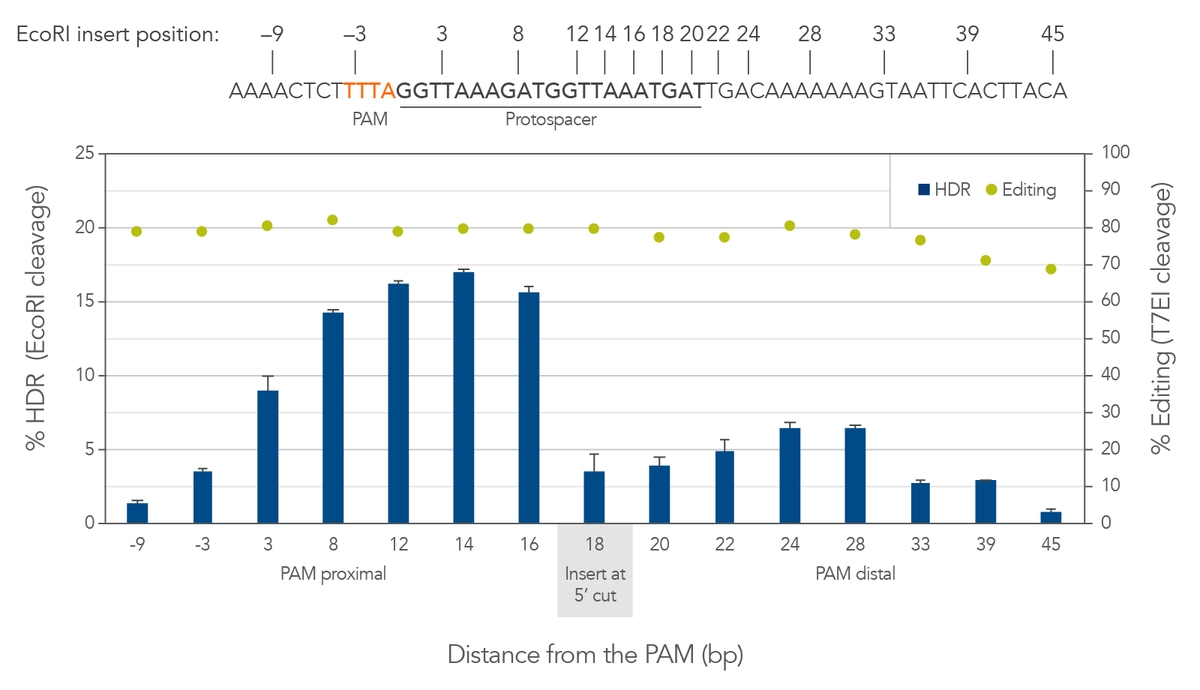
Cas12a creates a staggered double-stranded DNA cut. On the strand which contains the PAM, the cut is 18–19 bases away from the PAM in the 3′ direction (Figure 3, top). The cut on the opposite strand of the DNA is 23 bases from the PAM, resulting in a 5′ overhang of 4–5 bases. This is different from the blunt cut generated by Cas9. For Cas9, HDR is best facilitated by placing the insert directly at the dsDNA blunt cut site. For Cas12a we found that there is a better approach. We observed that walking the insert from the cut toward the PAM to disrupt the protospacer sequence can dramatically increase HDR by around 4-fold (Figure 3, bottom). Because Cas12a has tight binding affinity and specificity, disrupting the protospacer helps to prevent repeated rebinding and recutting by the Cas12a RNP at the locus where the insert is placed. The benefit of this shifted placement varies site-to-site; we recommend screening for each project to determine which combination of gRNAs and insert positions will result in optimal HDR.
Figure 3. For Cas12a HDR, walking the insert 2–6 bases from the cut site toward the PAM can dramatically increase HDR efficiency. HEK-293 cells were transfected by electroporation with wild-type A.s. Cas12a RNPs targeting human HPRT along with Ultramer HDR donor oligos containing a 6-base EcoRI insert and 40-nucleotide homology arms. Editing efficiency was measured by T7EI assay. HDR was measured by EcoRI cleavage of target amplicons; error bars represent the standard deviation of 3 technical replicates. Positioning the insert (the EcoRI site) 2–10 bases closer to the PAM increases the rate of HDR by fully disrupting the protospacer sequence, while positioning the insert further away from the cut site is disfavored. Optimizing the insert site location greatly improves the rate of HDR.
Use modified donors and Alt-R HDR Enhancer
To increase rates of HDR, we use donor oligos with stabilizing modifications, namely, 2 phosphorothioate (2PS) linkages at the 5′- and 3′-ends, to protect the donor oligos from exonuclease activity. Protecting the donor oligo from nuclease activity is especially impactful in high-nuclease environments such as iPS and Jurkat cells, which are notorious for poor HDR rates.
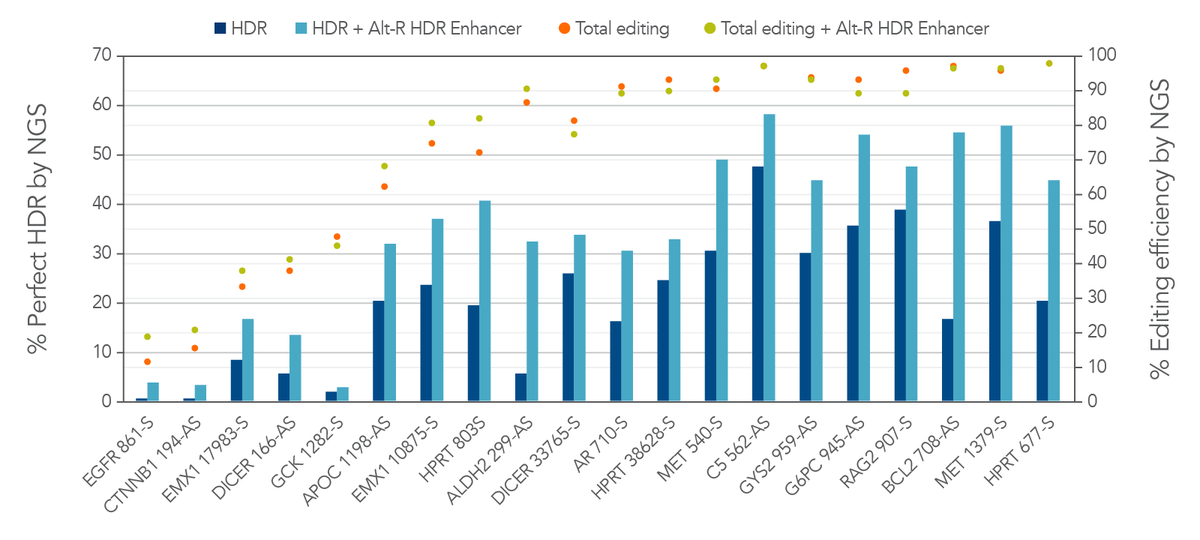
Alt-R HDR Enhancer further increases the rate of HDR by biasing the double-strand break repair pathways towards HDR rather than NHEJ. Figure 4 shows HDR rates using donor oligos containing the 2PS modifications in combination with Alt-R HDR Enhancer. HDR rates greater than 50% were observed in Jurkat cells. Therefore, HDR rates are greatly improved by adding appropriate chemical modifications to the donor and by using Alt-R HDR Enhancer.
Figure 4. 2PS-modified donor oligos and Alt-R HDR Enhancer can further improve the rate of HDR. Jurkat cells were transfected by electroporation with 2 µM Alt-R A.s. Cas12a Ultra RNPs targeting varied genomic loci along with 2PS-modified Ultramer HDR donor oligos containing a 6-base insert positioned 16 bases from the PAM and homology arms containing the non-targeted sequence. Cells were further treated with or without Alt-R HDR Enhancer. Editing efficiency and rate of perfect HDR were measured with NGS. By following the Cas12a Ultra HDR guidelines listed in this article, HDR rates >50% were observed in Jurkat cells, a cell line with typically low rates of HDR. HDR is increased with 2PS modifications of the donor and with Alt-R HDR Enhancer.
Summary
Together, these guidelines allow for high rates of HDR using the Alt-R A.s. Cas12a (Cpf1) Ultra system, even in challenging environments.
- Use a Cas12a nuclease with high activity like A.s. Cas12a Ultra, but use at a moderate concentration to avoid ssDNA degradation
- Use the non-targeted strand sequence for the ssDNA donor sequence
- Position your insert to disrupt the protospacer
- Use modified donors and Alt-R HDR Enhancer to boost the rate of HDR further
References
1. Chang HHY, Pannunzio NR, et al. (2017) Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat Rev Mol Cell Biol. 18(8):495–506.
2. Zetsche B, Gootenberg JS, et al. (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163(3):759–771.
3. Kim D, Kim J, et al. (2016) Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol. 34(8):863–868.
4. Tsai SQ, Zheng Z, et al. (2015) GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 33(2):187–197.
(c) 2019 by Bernice Thommandru, MS, Research Scientist, IDT